7T4V
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-methylbenzoic acid, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4W
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | (2R)-2-({(2S,3S)-1-[(1H-benzimidazol-2-yl)methyl]-2-phenylpiperidin-3-yl}oxy)-2-(3,5-dichlorophenyl)ethan-1-ol, CHLORIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
7T4T
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
5YLT
 
 | | Crystal structure of SET7/9 in complex with a cyproheptadine derivative | | Descriptor: | 2-(1-methylpiperidin-4-ylidene)tricyclo[9.4.0.0^{3,8}]pentadeca-1(11),3(8),4,6,9,12,14-heptaen-6-ol, GLYCEROL, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Hirano, T, Fujiwara, T, Niwa, H, Hirano, M, Ohira, K, Okazaki, Y, Sato, S, Umehara, T, Maemoto, Y, Ito, A, Yoshida, M, Kagechika, H. | | Deposit date: | 2017-10-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of Novel Inhibitors for Histone Methyltransferase SET7/9 based on Cyproheptadine.
ChemMedChem, 13, 2018
|
|
7T4U
 
 | | Crystal Structure of cGMP-dependent Protein Kinase | | Descriptor: | 3-amino-4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)benzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Zebisch, M, Silvestre, L, Fischmann, T.O. | | Deposit date: | 2021-12-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Selective small molecule activation of PKG1alpha: structure and function
Commun Biol, 2023
|
|
5UHY
 
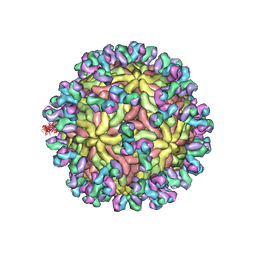 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | Descriptor: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | Authors: | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
5BYE
 
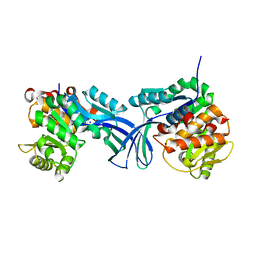 | | Crystal structure of human ribokinase in P212121 spacegroup | | Descriptor: | CHLORIDE ION, Ribokinase, SODIUM ION | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of human ribokinase in P212121 spacegroup
To Be Published
|
|
1KI8
 
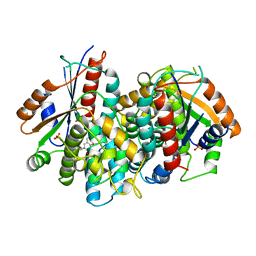 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH 5-BROMOVINYLDEOXYURIDINE | | Descriptor: | 5-BROMOVINYLDEOXYURIDINE, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Visse, R, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
3ONF
 
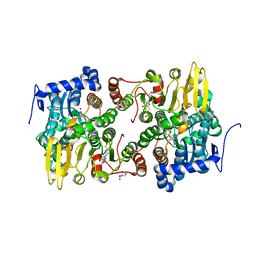 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4QTI
 
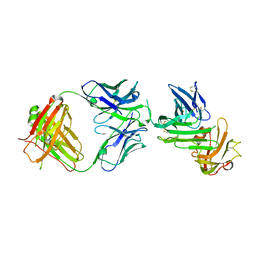 | | Crystal structure of human uPAR in complex with anti-uPAR Fab 8B12 | | Descriptor: | Urokinase plasminogen activator surface receptor, anti-uPAR antibody, heavy chain, ... | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
7OTL
 
 | | Structure of a psychrophilic CCA-adding enzyme crystallized by counter-diffusion | | Descriptor: | CCA-adding enzyme, SULFATE ION | | Authors: | Rollet, K, de Wijn, R, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2021-06-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
7ORZ
 
 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-06 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OQX
 
 | | Crystal structure of a psychrophilic CCA-adding enzyme in complex with CMPcPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ACETATE ION, CCA-adding enzyme, ... | | Authors: | Rollet, K, de Wijn, R, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
7OTR
 
 | | Crystal structure of a psychrophilic CCA-adding enzyme determined by SAD phasing | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CCA-adding protein, ... | | Authors: | Rollet, K, de Wijn, R, Rios-Santacruz, R, Hennig, O, Betat, H, Moerl, M, Sauter, C. | | Deposit date: | 2021-06-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus .
Comput Struct Biotechnol J, 19, 2021
|
|
5CA7
 
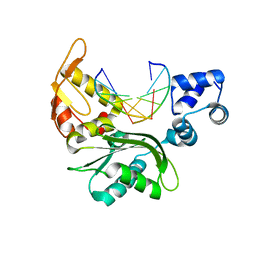 | |
3P0C
 
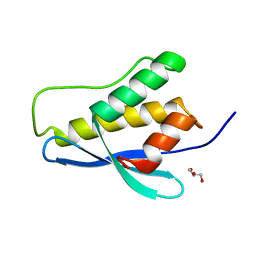 | | Nischarin PX-domain | | Descriptor: | GLYCEROL, Nischarin | | Authors: | Schutz, P, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Der Berg, S, Wahlberg, E, Welin, M, Weigelt, J, Nordlund, P, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-28 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Crystal structure of Nischarin PX-domain
to be published
|
|
4RBW
 
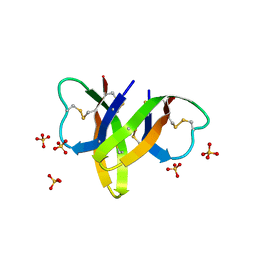 | | Crystal structure of human alpha-defensin 5, HD5 (Thr7Arg Glu21Arg mutant) | | Descriptor: | CHLORIDE ION, Defensin-5, SULFATE ION | | Authors: | Pazgier, M, Gohain, N, Tolbert, W.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a potent antibiotic peptide based on the active region of human defensin 5.
J.Med.Chem., 58, 2015
|
|
5C2W
 
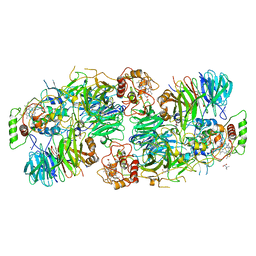 | | Kuenenia stuttgartiensis Hydrazine Synthase Pressurized with 20 bar Xenon | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEME C, ... | | Authors: | Dietl, A, Ferousi, C, Maalcke, W.J, Menzel, A, de Vries, S, Keltjens, J.T, Jetten, M.S.M, Kartal, B, Barends, T.R.M. | | Deposit date: | 2015-06-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The inner workings of the hydrazine synthase multiprotein complex.
Nature, 527, 2015
|
|
5BYC
 
 | | Crystal structure of human ribokinase in C2 spacegroup | | Descriptor: | Ribokinase, SODIUM ION | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of unliganded human ribokinase in C2 spacegroup
To Be Published
|
|
4RAX
 
 | | A regulatory domain of an ion channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Ge, J, Yang, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Architecture of the mammalian mechanosensitive Piezo1 channel.
Nature, 527, 2015
|
|
5C41
 
 | | Crystal structure of human ribokinase in complex with AMPPCP in P21 spacegroup and with 4 protomers | | Descriptor: | PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ribokinase, ... | | Authors: | Park, J, Chakrabarti, J, Singh, B, Gupta, R.S, Junop, M.S. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human ribokinase in complex with AMPPCP in P21 spacegroup and with 4 protomers
To Be Published
|
|
6RY7
 
 | |
5CB1
 
 | | Apo enzyme of human Polymerase lambda | | Descriptor: | DNA polymerase lambda | | Authors: | Liu, M.S, Tsai, M.D. | | Deposit date: | 2015-06-30 | | Release date: | 2016-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Mechanism for the Fidelity Modulation of DNA Polymerase lambda
J.Am.Chem.Soc., 138, 2016
|
|
5YGX
 
 | | Structure of BACE1 in complex with N-(3-((4R,5R,6S)-2-amino-6-(1,1-difluoroethyl)-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Nakahara, K, Fuchino, K, Komano, K, Asada, N, Tadano, G, Hasegawa, T, Yamamoto, T, Sako, Y, Ogawa, M, Unemura, C, Hosono, M, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | Deposit date: | 2017-09-27 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Potent and Centrally Active 6-Substituted 5-Fluoro-1,3-dihydro-oxazine beta-Secretase (BACE1) Inhibitors via Active Conformation Stabilization
J. Med. Chem., 61, 2018
|
|
4R8U
 
 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
