2ZHL
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 2) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZG8
 
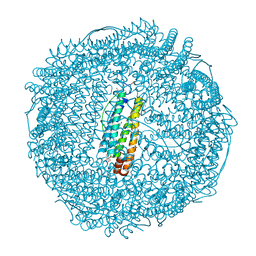 | | Crystal Structure of Pd(allyl)/apo-H49AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Niemeyer, J, Abe, M, Ueno, T, Hikage, T, Erker, G, Watanabe, Y. | | Deposit date: | 2008-01-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Control of the coordination structure of organometallic palladium complexes in an apo-ferritin cage.
J.Am.Chem.Soc., 130, 2008
|
|
2ZHK
 
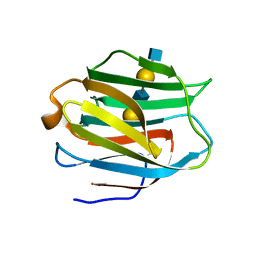 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer (crystal 1) | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
3UKW
 
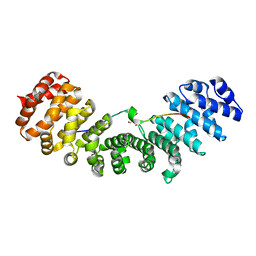 | | Mouse importin alpha: Bimax1 peptide complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bimax1 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
2LIP
 
 | |
2ZJX
 
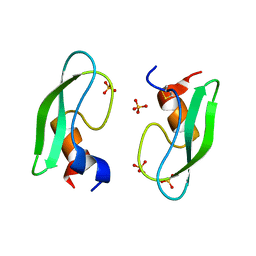 | | Bovine pancreatic trypsin inhibitor (BPTI) containing only the [5,55] disulfide bond | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Sohya, S, Noguchi, K, Yohda, M, Kuroda, Y. | | Deposit date: | 2008-03-11 | | Release date: | 2008-10-21 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of an extensively simplified variant of bovine pancreatic trypsin inhibitor in which over one-third of the residues are alanines
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4E82
 
 | |
1E94
 
 | | HslV-HslU from E.coli | | Descriptor: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | Deposit date: | 2000-10-07 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ZJL
 
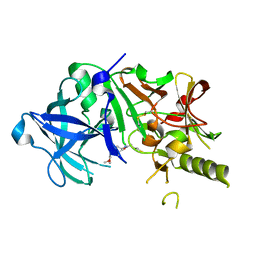 | | Crystal structure of the human BACE1 catalytic domain in complex with N-[1-(5-bromo-2,3-dimethoxy-benzyl)-piperidin-4-yl]-4-mercapto-butyramide | | Descriptor: | Beta-secretase 1, N-[1-(5-bromo-2,3-dimethoxybenzyl)piperidin-4-yl]-4-sulfanylbutanamide | | Authors: | Randal, M, Lam, M.B, Lu, W, Romanowski, M.J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based discovery of novel BACE1 inhibitors using Tethering technology
To be Published
|
|
2ZMA
 
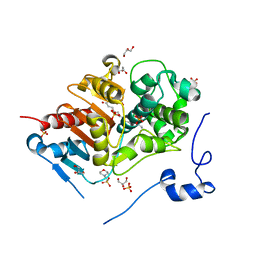 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
3HHY
 
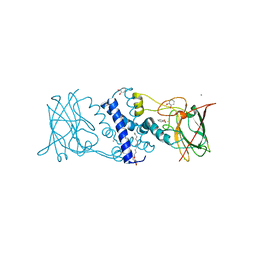 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with catechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CATECHOL, CHLORIDE ION, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-05-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3UDP
 
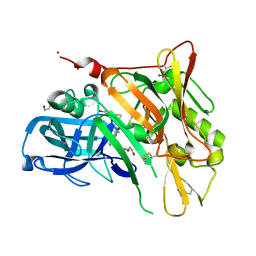 | | Crystal Structure of BACE with Compound 12 | | Descriptor: | (4S)-6-bromo-3,4-dihydro-2H-thiochromen-4-yl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
2Z29
 
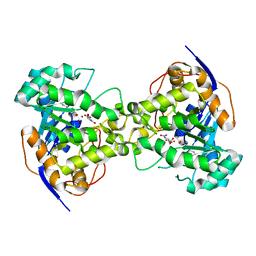 | | Thr109Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
3UJ0
 
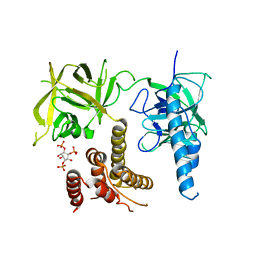 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor with ligand bound form. | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
2LUG
 
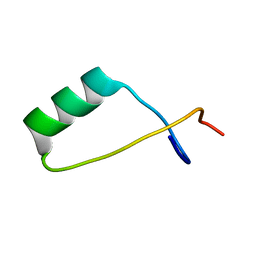 | | Solution NMR structure of a S72-S107 peptide of 18.5kDa murine myelin basic protein (MBP) in association with dodecylphosphocholine micelles | | Descriptor: | Myelin basic protein | | Authors: | Ahmed, M.A.M, De Avila, M, Polverini, E, Bessonov, K, Bamm, V.V, Harauz, G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Nuclear Magnetic Resonance Structure and Molecular Dynamics Simulations of a Murine 18.5 kDa Myelin Basic Protein Segment (S72-S107) in Association with Dodecylphosphocholine Micelles.
Biochemistry, 51, 2012
|
|
2Z33
 
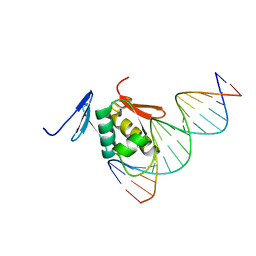 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | Descriptor: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
4EFX
 
 | |
2LUJ
 
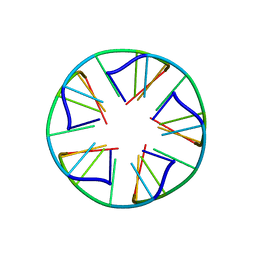 | | Solution structure of a parallel-stranded oligoisoguanine DNA pentaplex formed by d(T(iG)4T) in the presence of Cs ions | | Descriptor: | DNA (5'-D(*TP*(IGU)P*(IGU)P*(IGU)P*(IGU)P*T)-3') | | Authors: | Kang, M, Heuberger, B, Chaput, J.C, Switzer, C, Feigon, J. | | Deposit date: | 2012-06-14 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Parallel-Stranded Oligoisoguanine DNA Pentaplex Formed by d(T(iG)4T) in the Presence of Cs(+) Ions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2MKS
 
 | |
2Z5I
 
 | | Crystal structure of the head-to-tail junction of tropomyosin | | Descriptor: | General control protein GCN4 and Tropomyosin alpha-1 chain, MAGNESIUM ION, Tropomyosin alpha-1 chain and General control protein GCN4 | | Authors: | Murakami, K, Nozawa, K, Tomii, K, Kudou, N, Igarashi, N, Shirakihara, Y, Wakatsuki, S, Stewart, M, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 2007-07-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2ZPL
 
 | | Crystal structure analysis of PDZ domain A | | Descriptor: | GLYCEROL, NICKEL (II) ION, Regulator of sigma E protease | | Authors: | Inaba, K, Suzuki, M. | | Deposit date: | 2008-07-17 | | Release date: | 2008-10-21 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of PDZ-domain A
To be Published
|
|
4Y0R
 
 | | Bovine beta-lactoglobulin complex with pramocaine crystallized from ammonium sulphate (BLG-PRM2) | | Descriptor: | Beta-lactoglobulin, Pramocaine | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
2Z5V
 
 | | Solution structure of the TIR domain of human MyD88 | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Ohnishi, H, Tochio, H, Hiroaki, H, Kondo, N, Kato, Z, Shirakawa, M. | | Deposit date: | 2007-07-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling.
Proc.Natl.Acad.Sci.USA, 2009
|
|
3HEI
 
 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
2ZR0
 
 | | MSRECA-Q196E mutant | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein recA | | Authors: | Prabu, J.R, Manjunath, G.P, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functionally important movements in RecA molecules and filaments: studies involving mutation and environmental changes
Acta Crystallogr.,Sect.D, 64, 2008
|
|
