1SMP
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN SERRATIA MARCESCENS METALLO-PROTEASE AND AN INHIBITOR FROM ERWINIA CHRYSANTHEMI | | Descriptor: | CALCIUM ION, ERWINIA CHRYSANTHEMI INHIBITOR, SERRATIA METALLO PROTEINASE, ... | | Authors: | Baumann, U, Bauer, M, Letoffe, S, Delepelaire, P, Wandersman, C. | | Deposit date: | 1995-01-13 | | Release date: | 1996-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi.
J.Mol.Biol., 248, 1995
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1S9A
 
 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
1SKV
 
 | | Crystal Structure of D-63 from Sulfolobus Spindle Virus 1 | | Descriptor: | Hypothetical 7.5 kDa protein | | Authors: | Kraft, P, Kummel, D, Oeckinghaus, A, Gauss, G.H, Wiedenheft, B, Young, M, Lawrence, C.M. | | Deposit date: | 2004-03-05 | | Release date: | 2004-07-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of d-63 from sulfolobus spindle-shaped virus 1: surface properties of the dimeric four-helix bundle suggest an adaptor protein function
J.Virol., 78, 2004
|
|
7KOD
 
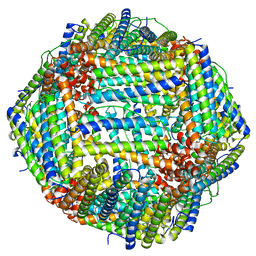 | | Cryo-EM structure of heavy chain mouse apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Sun, M, Azumaya, C, Tse, E, Frost, A, Southworth, D, Verba, K.A, Cheng, Y, Agard, D.A. | | Deposit date: | 2020-11-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.655 Å) | | Cite: | Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM.
J.Struct.Biol., 213, 2021
|
|
1SEH
 
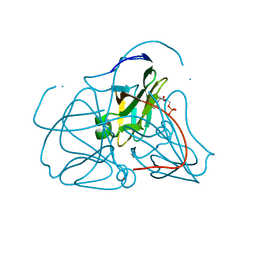 | | Crystal structure of E. coli dUTPase complexed with the product dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-02-17 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
4FEZ
 
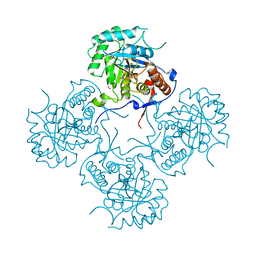 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant.
To be Published
|
|
1SHV
 
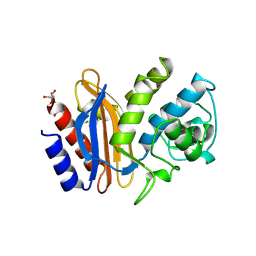 | | STRUCTURE OF SHV-1 BETA-LACTAMASE | | Descriptor: | CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, PROTEIN (BETA-LACTAMASE SHV-1) | | Authors: | Kuzin, A.P, Nukaga, M, Nukaga, Y, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 1999-02-23 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the SHV-1 beta-lactamase.
Biochemistry, 38, 1999
|
|
1SIF
 
 | | Crystal structure of a multiple hydrophobic core mutant of ubiquitin | | Descriptor: | ubiquitin | | Authors: | Benitez-Cardoza, C.G, Stott, K, Hirshberg, M, Went, H.M, Woolfson, D.N, Jackson, S.E. | | Deposit date: | 2004-02-29 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Exploring sequence/folding space: folding studies on multiple hydrophobic core mutants of ubiquitin
Biochemistry, 43, 2004
|
|
1SIQ
 
 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
1SMQ
 
 | | Structure of the Ribonucleotide Reductase Rnr2 Homodimer from Saccharomyces cerevisiae | | Descriptor: | Ribonucleoside-diphosphate reductase small chain 1 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
2WNS
 
 | | Human Orotate phosphoribosyltransferase (OPRTase) domain of Uridine 5' -monophosphate synthase (UMPS) in complex with its substrate orotidine 5'-monophosphate (OMP) | | Descriptor: | CHLORIDE ION, OROTATE PHOSPHORIBOSYLTRANSFERASE, OROTIDINE-5'-MONOPHOSPHATE | | Authors: | Moche, M, Roos, A, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotyenova, T, Kotzch, A, Nielsen, T.K, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, VanDenBerg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-07-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Orotate Phosphoribosyltransferase (Oprtase) Domain of Uridine 5'-Monophosphate Synthase (Umps) in Complex with its Substrate Orotidine 5'-Monophosphate (Omp)
To be Published
|
|
1SOT
 
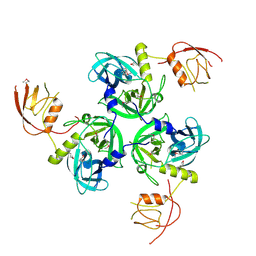 | | Crystal Structure of the DegS stress sensor | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
1SJ9
 
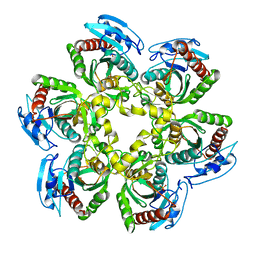 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium at 2.5A resolution | | Descriptor: | PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M, Gabdoulkhakov, A, Morgunova, E, Garber, M, Nikonov, S, Betzel, C, Ealick, S, Mikhailov, A. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4H03
 
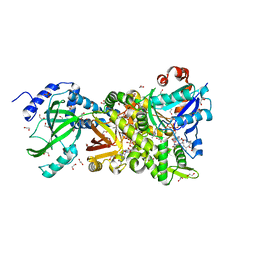 | | Crystal structure of NAD+-Ia-actin complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Tsurumura, T, Oda, M, Nagahama, M, Tsuge, H. | | Deposit date: | 2012-09-07 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arginine ADP-ribosylation mechanism based on structural snapshots of iota-toxin and actin complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HGD
 
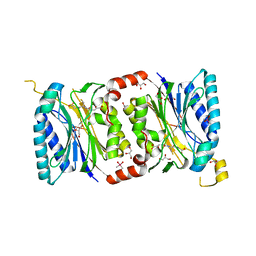 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1S88
 
 | | NMR structure of a DNA duplex with two INA nucleotides inserted opposite each other, dCTCAACXCAAGCT:dAGCTTGXGTTGAG | | Descriptor: | 5'-D(*AP*GP*CP*TP*TP*GP*(2DM)P*GP*TP*TP*GP*AP*G)-3', 5'-D(*CP*TP*CP*AP*AP*CP*(2DM)P*CP*AP*AP*GP*CP*T)-3' | | Authors: | Nielsen, C.B, Petersen, M, Pedersen, E.B, Hansen, P.E, Christensen, U.B. | | Deposit date: | 2004-01-31 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid.
Bioconjug.Chem., 15, 2004
|
|
5AAY
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | NF-KAPPA-B ESSENTIAL MODULATOR, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
1SXJ
 
 | | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Activator 1 37 kDa subunit, Activator 1 40 kDa subunit, ... | | Authors: | Bowman, G.D, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.
Nature, 429, 2004
|
|
1SX0
 
 | | Solution NMR Structure and X-Ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase | | Descriptor: | SecA | | Authors: | Dempsey, B.R, Wrona, M, Moulin, J.M, Gloor, G.B, Jalilehvand, F, Lajoie, G, Shaw, G.S, Shilton, B.H. | | Deposit date: | 2004-03-30 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and X-ray Absorption Analysis of the C-Terminal Zinc-Binding Domain of the SecA ATPase.
Biochemistry, 43, 2004
|
|
1SXE
 
 | | The solution structure of the Pointed (PNT) domain from the transcrition factor Erg | | Descriptor: | Transcriptional regulator ERG | | Authors: | Mackereth, C.D, Schaerpf, M, Gentile, L.N, MacIntosh, S.E, Slupsky, C.M, McIntosh, L.P. | | Deposit date: | 2004-03-30 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Diversity in Structure and Function of the Ets Family PNT Domains.
J.Mol.Biol., 342, 2004
|
|
1SYL
 
 | | Crystal structure of inactive mutant dUTPase complexed with substrate dUTP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE-5'-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Barabas, O, Kovari, J, Pongracz, V, Wilmanns, M, Vertessy, B.G. | | Deposit date: | 2004-04-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase
J.Biol.Chem., 279, 2004
|
|
1T12
 
 | | Solution Structure of a new LTP1 | | Descriptor: | NONSPECIFIC LIPID-TRANSFER PROTEIN 1 | | Authors: | da Silva, P, Landon, C, Industri, B, Ponchet, M, Vovelle, F. | | Deposit date: | 2004-04-15 | | Release date: | 2005-04-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tobacco lipid transfer protein exhibiting
new biophysical and biological features
Proteins, 59, 2005
|
|
