5S24
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-697611 | | Descriptor: | 2-(1H-benzimidazol-1-yl)-N-methylacetamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1JPX
 
 | | Mutation that destabilize the gp41 core: determinants for stabilizing the SIV/CPmac envelope glycoprotein complex. Wild type. | | Descriptor: | gp41 envelope protein | | Authors: | Liu, J, Wang, S, LaBranche, C.C, Hoxie, J.A, Lu, M. | | Deposit date: | 2001-08-03 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations that destabilize the gp41 core are determinants for stabilizing the simian immunodeficiency virus-CPmac envelope glycoprotein complex.
J.Biol.Chem., 277, 2002
|
|
5S2N
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1787627869 | | Descriptor: | 5-chloranyl-~{N}-methyl-~{N}-[[(3~{S})-oxolan-3-yl]methyl]pyrimidin-4-amine, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.133 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4QGW
 
 | | Crystal sturcture of the R132K:R111L:L121D mutant of Cellular Retinoic Acid Binding ProteinII complexed with a synthetic ligand (Merocyanine) at 1.77 angstrom resolution | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Nosrati, M, Yapici, I, Geiger, J.H. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
5S1O
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with STL414928 | | Descriptor: | 2H-pyrazolo[3,4-b]pyridin-5-amine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4HS6
 
 | |
5S32
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z26781943 | | Descriptor: | N-[(1H-benzimidazol-2-yl)methyl]-2-methylpropanamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.166 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4Q4J
 
 | | Structure of crosslinked TM287/288_S498C_S520C mutant | | Descriptor: | ABC transporter, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Hohl, M, Schoeppe, J, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5S2C
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z45612755 | | Descriptor: | N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)methanesulfonamide, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.092 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4QH4
 
 | |
5S3I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z50145861 | | Descriptor: | 5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6K2R
 
 | |
3FYR
 
 | | Crystal structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis | | Descriptor: | Sporulation inhibitor sda | | Authors: | Jacques, D.A, Streamer, M, King, G.F, Guss, J.M, Trewhella, J, Langley, D.B. | | Deposit date: | 2009-01-23 | | Release date: | 2009-06-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4Q6M
 
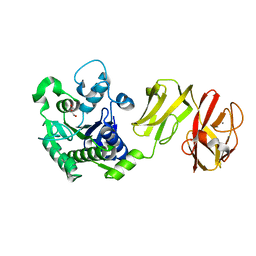 | | Structural analysis of the apo-form of Helicobacter pylori Csd4, a D,L-carboxypeptidase | | Descriptor: | CALCIUM ION, Conserved hypothetical secreted protein, GLYCEROL | | Authors: | Kim, H.S, Kim, J, Im, H.N, An, D.R, Lee, M, Hesek, D, Mobashery, S, Kim, J.Y, Cho, K, Yoon, H.J, Han, B.W, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-04-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6K31
 
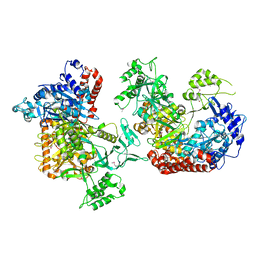 | |
3FZ0
 
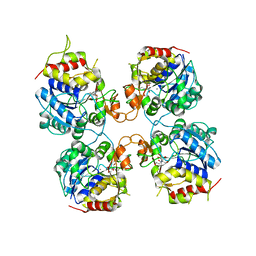 | | Inosine-Guanosine Nucleoside Hydrolase (IG-NH) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Nucleoside hydrolase, ... | | Authors: | Vandemeulebroucke, A, Minici, C, Bruno, I, Muzzolini, L, Tornaghi, P, Parkin, D.W, Schramm, V.L, Versees, W, Steyaert, J, Degano, M. | | Deposit date: | 2009-01-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the 6-oxopurine nucleosidase from Trypanosoma brucei brucei
Biochemistry, 49, 2010
|
|
4KEU
 
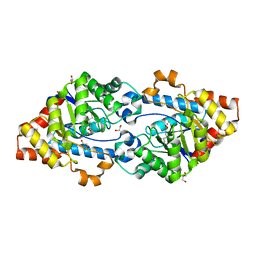 | | Crystal structure of SsoPox W263M | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
3RW7
 
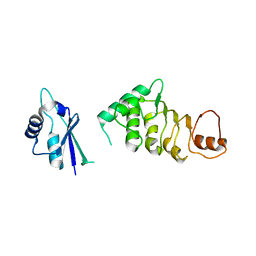 | | Structure of N-terminal domain of nuclear RNA export factor TAP | | Descriptor: | Nuclear RNA export factor 1 | | Authors: | Teplova, M, Khin, N.W, Wohlbold, L, Izaurralde, E, Patel, D.J. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3VJH
 
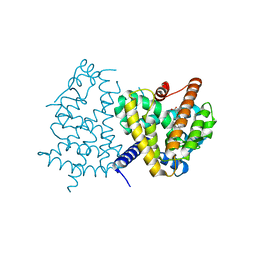 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | Descriptor: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
4PWG
 
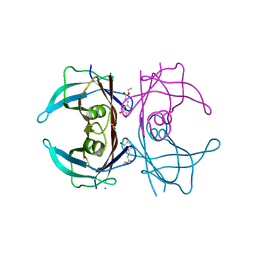 | | Crystal structure of V30M mutant human transthyretin complexed with caffeic acid ethyl ester | | Descriptor: | CALCIUM ION, Transthyretin, ethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate | | Authors: | Yokoyama, T, Kosaka, Y, Mizuguchi, M. | | Deposit date: | 2014-03-20 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Inhibitory Activities of Propolis and Its Promising Component, Caffeic Acid Phenethyl Ester, against Amyloidogenesis of Human Transthyretin
J.Med.Chem., 57, 2014
|
|
3S9G
 
 | |
1JWK
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457A Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
4Q7K
 
 | |
3RXA
 
 | | Crystal structure of Trypsin complexed with cycloheptanamine | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
4Q7L
 
 | | Structure of NBD288 of TM287/288 | | Descriptor: | NICKEL (II) ION, Uncharacterized ABC transporter ATP-binding protein TM_0288 | | Authors: | Bukowska, M.A, Hohl, M, Gruetter, M.G, Seeger, M.A. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Transporter Motor Taken Apart: Flexibility in the Nucleotide Binding Domains of a Heterodimeric ABC Exporter.
Biochemistry, 54, 2015
|
|
