6GV9
 
 | | Structure of the type IV pilus from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Bardiaux, B, Amorim, G.C, Luna-Rico, A, Zheng, W, Guilvout, I, Jollivet, C, Nilges, M, Egelman, E, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
4O3T
 
 | | Zymogen HGF-beta/MET with Zymogen Activator Peptide ZAP.14 | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor, PENTAETHYLENE GLYCOL, ... | | Authors: | Eigenbrot, C, Landgraf, K.E, Steffek, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | An allosteric switch for pro-HGF/Met signaling using zymogen activator peptides.
Nat.Chem.Biol., 10, 2014
|
|
3OVW
 
 | | ENDOGLUCANASE I NATIVE STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Schulein, M. | | Deposit date: | 1997-10-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the endoglucanase I from Fusarium oxysporum: native, cellobiose, and 3,4-epoxybutyl beta-D-cellobioside-inhibited forms, at 2.3 A resolution.
Biochemistry, 36, 1997
|
|
1JXQ
 
 | | Structure of cleaved, CARD domain deleted Caspase-9 | | Descriptor: | Caspase-9, benzoxycarbonyl-Val-Ala-Asp-fluoromethyl ketone Inhibitor | | Authors: | Renatus, M, Stennicke, H.R, Scott, F.L, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2001-09-08 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer formation drives the activation of the cell death protease caspase 9.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4O4Q
 
 | | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the complex formed between type 1 ribosome inactivating protein and uridine diphosphate at 1.81 A resolution
To be Published
|
|
6GPJ
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
4O4L
 
 | | Tubulin-Peloruside A-Epothilone A complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, EPOTHILONE A, ... | | Authors: | Prota, A.E, Bargsten, K, Northcote, P.T, Marsh, M, Altmann, K.H, Miller, J.H, Diaz, J.F, Steinmetz, M.O. | | Deposit date: | 2013-12-18 | | Release date: | 2014-03-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of microtubule stabilization by laulimalide and peloruside a.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6HT4
 
 | |
2Y6E
 
 | | Structure of the D1D2 domain of USP4, the conserved catalytic domain | | Descriptor: | SULFATE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 4, ZINC ION | | Authors: | Luna-Vargas, M.P.A, Faesen, A.C, van Dijk, W.J, Rape, M, Fish, A, Sixma, T.K. | | Deposit date: | 2011-01-20 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Dusp-Ubl Domain of Usp4 Enhances its Catalytic Efficiency by Promoting Ubiquitin Exchange.
Nat.Commun., 5, 2014
|
|
4O55
 
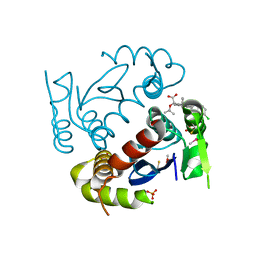 | |
6GWQ
 
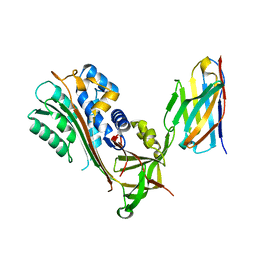 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-stab) in Complex with an Inhibitory Nanobody (VHH-2g-42) | | Descriptor: | Plasminogen Activator Inhibitor-1, VHH-2g-42 | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2018-06-25 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
6HYH
 
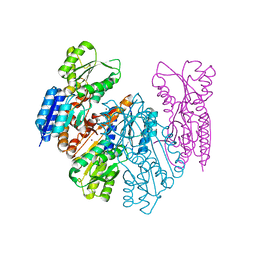 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
4O5A
 
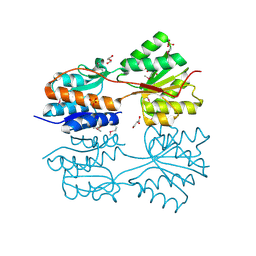 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
3DL9
 
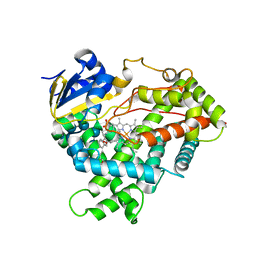 | | Crystal structure of CYP2R1 in complex with 1-alpha-hydroxy-vitamin D2 | | Descriptor: | (1S,3R,5Z,7E,22E)-9,10-secoergosta-5,7,10,22-tetraene-1,3-diol, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cytochrome P450 2R1, ... | | Authors: | Strushkevich, N.V, Tempel, W, Gilep, A.A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Wilkstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.721 Å) | | Cite: | Crystal structure of CYP2R1 in complex with 1-alpha-hydroxy-vitamin D2.
To be Published
|
|
4KSB
 
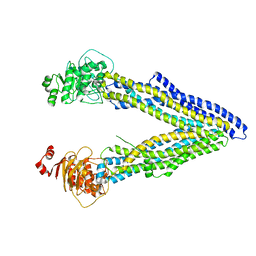 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1JHT
 
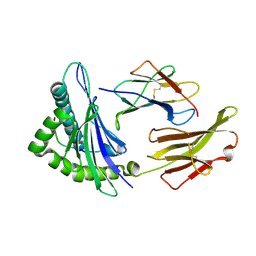 | | Crystal structure of HLA-A2*0201 in complex with a nonameric altered peptide ligand (ALGIGILTV) from the MART-1/Melan-A. | | Descriptor: | HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, beta-2-microglobulin, ... | | Authors: | Sliz, P, Michielin, O, Karplus, M, Romero, P, Wiley, D. | | Deposit date: | 2001-06-28 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of two closely related but antigenically distinct HLA-A2/melanocyte-melanoma tumor-antigen peptide complexes.
J.Immunol., 167, 2001
|
|
4O4O
 
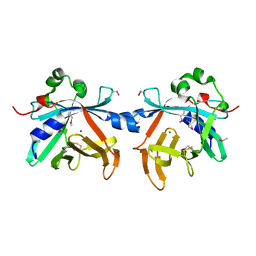 | | Crystal structure of phycobiliprotein lyase CpcT | | Descriptor: | MAGNESIUM ION, Phycocyanobilin lyase CpcT | | Authors: | Zhou, W, Ding, W.-L, Zeng, X.-l, Dong, L.-L, Zhao, B, Zhou, M, Scheer, H, Zhao, K.-H, Yang, X. | | Deposit date: | 2013-12-19 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of the Phycobiliprotein Lyase CpcT.
J.Biol.Chem., 289, 2014
|
|
6HYW
 
 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT Y119F | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1K0N
 
 | | Chloride Intracellular Channel 1 (CLIC1) complexed with glutathione | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1, GLUTATHIONE | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
1JI7
 
 | | Crystal Structure of TEL SAM Polymer | | Descriptor: | ETS-RELATED PROTEIN TEL1, SULFATE ION | | Authors: | Kim, C.A, Phillips, M.L, Kim, W, Gingery, M, Tran, H.H, Robinson, M.A, Faham, S, Bowie, J.U. | | Deposit date: | 2001-06-29 | | Release date: | 2002-07-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression.
EMBO J., 20, 2001
|
|
3P09
 
 | | Crystal Structure of Beta-Lactamase from Francisella tularensis | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Beta-Lactamase from Francisella tularensis
To be Published
|
|
6I0S
 
 | | Crystal structure of DmTailor in complex with UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, Terminal uridylyltransferase Tailor | | Authors: | Kroupova, A, Ivascu, A, Jinek, M. | | Deposit date: | 2018-10-26 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
6GFF
 
 | | Structure of GARP (LRRC32) in complex with latent TGF-beta1 and MHG-8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat-containing protein 32, MHG-8 Fab heavy chain, ... | | Authors: | Merceron, R, Lienart, S, Vanderaa, C, Colau, D, Stockis, J, Van Der Woning, B, De Haard, H, Saunders, M, Coulie, P.G, Savvides, S.N, Lucas, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of latent TGF-beta 1 presentation and activation by GARP on human regulatory T cells.
Science, 362, 2018
|
|
3P1A
 
 | | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Chaikuad, A, Eswaran, J, Fedorov, O, Cooper, C.D.O, Kroeler, T, Vollmar, M, Krojer, T, Berridge, G, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1)
To be Published
|
|
6TK3
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 30us+150us structure of KR2 with extrapolated, light and dark datasets | | Descriptor: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | Authors: | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
