6JUI
 
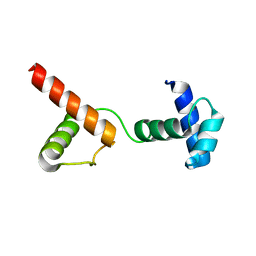 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
6KIQ
 
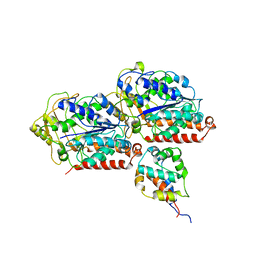 | | Complex of yeast cytoplasmic dynein MTBD-High and MT with DTT | | Descriptor: | Alpha tubulin, Dynein heavy chain, cytoplasmic, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
4FKB
 
 | |
6KJN
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the high affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6K7M
 
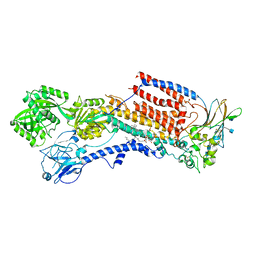 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2Pi-PL state) | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
4FML
 
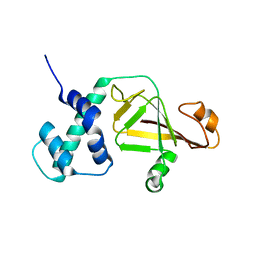 | |
6K4Z
 
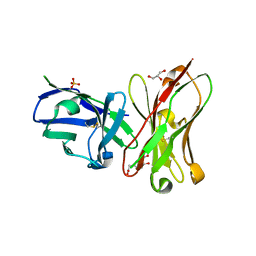 | | Single-chain Fv antibody of C6 COMPLEXED WITH 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID | | Descriptor: | 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Nishiguchi, A, Numoto, N, Ito, N, Azuma, T, Oda, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-dimensional structure of a high affinity anti-(4-hydroxy-3-nitrophenyl)acetyl antibody possessing a glycine residue at position 95 of the heavy chain.
Mol.Immunol., 114, 2019
|
|
7Z6R
 
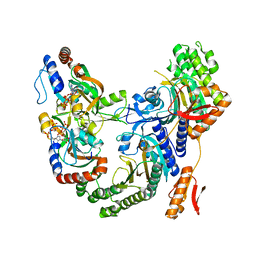 | | Psychrobacter arcticus ATPPRT (HisGZ) R56A mutant bound to ATP and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Fisher, G, da Silva, R.G. | | Deposit date: | 2022-03-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric rescue of catalytically impaired ATP phosphoribosyltransferase variants links protein dynamics to active-site electrostatic preorganisation.
Nat Commun, 13, 2022
|
|
6K9L
 
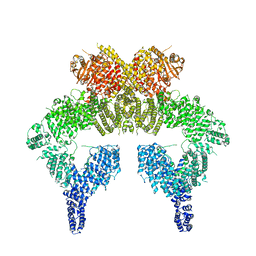 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
6KCR
 
 | |
7Z8U
 
 | | Catalytic subunit HisG R56A mutant from Psychrobacter arcticus ATPPRT (HisGZ) in complex with ATP and PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Fisher, G, da Silva, R.G. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric rescue of catalytically impaired ATP phosphoribosyltransferase variants links protein dynamics to active-site electrostatic preorganisation.
Nat Commun, 13, 2022
|
|
6KFF
 
 | | Undocked INX-6 hemichannel in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KFH
 
 | | Undocked hemichannel of an N-terminal deletion mutant of INX-6 in a nanodisc | | Descriptor: | Innexin-6 | | Authors: | Burendei, B, Shinozaki, R, Watanabe, M, Terada, T, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2019-07-07 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of undocked innexin-6 hemichannels in phospholipids.
Sci Adv, 6, 2020
|
|
6KKA
 
 | | Xylanase J mutant from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, M, Takita, T, Nakatani, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6D7J
 
 | |
4FMP
 
 | | Crystal structure of thermostable, organic-solvent tolerant lipase from Geobacillus sp. strain ARM | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Nisbar, N.D, Rahman, R.N.Z.R.A, Ali, M.S.M, Leow, A.T.C. | | Deposit date: | 2012-06-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of novel ARM lipase and elucidation of its space-grown crystal structure
Thesis, 2013
|
|
6KFN
 
 | | Crystal structure of alginate lyase from Paenibacillus sp. str. FPU-7 | | Descriptor: | IMIDAZOLE, SODIUM ION, alginate lyase | | Authors: | Itoh, T, Nakagawa, E, Yoda, M, Nakaichi, A, Hibi, T, Kimoto, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7.
Sci Rep, 9, 2019
|
|
6KG6
 
 | | Crystal structure of MavC/UBE2N-Ub complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wang, Y, Huang, Y, Chang, M, Feng, Y. | | Deposit date: | 2019-07-10 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
6KML
 
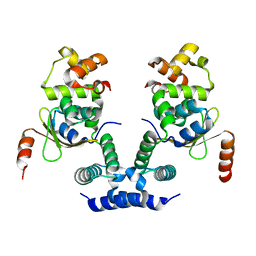 | | 2.09 Angstrom resolution crystal structure of tetrameric HigBA toxin-antitoxin complex from E.coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KMQ
 
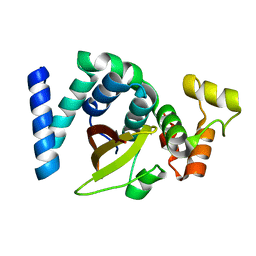 | | 2.3 Angstrom resolution structure of dimeric HigBA toxin-antitoxin complex from E. coli | | Descriptor: | Antitoxin HigA, mRNA interferase toxin HigB | | Authors: | Jadhav, P, Sinha, V.K, Rothweiler, U, Singh, M. | | Deposit date: | 2019-07-31 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.09 angstrom Resolution structure of E. coli HigBA toxin-antitoxin complex reveals an ordered DNA-binding domain and intrinsic dynamics in antitoxin.
Biochem.J., 477, 2020
|
|
6KIO
 
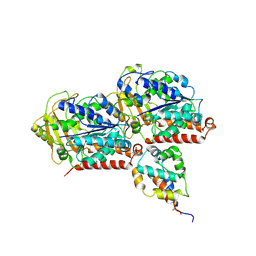 | | Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT | | Descriptor: | Dynein heavy chain, cytoplasmic, Tubulin alpha-1A chain, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6L7A
 
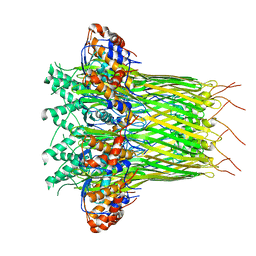 | | CsgFG complex in Curli biogenesis system | | Descriptor: | CsgF, Curli production assembly/transport protein CsgG | | Authors: | Yan, Z.F, Yin, M, Chen, J.N, Li, X.M. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Assembly and substrate recognition of curli biogenesis system.
Nat Commun, 11, 2020
|
|
6KRE
 
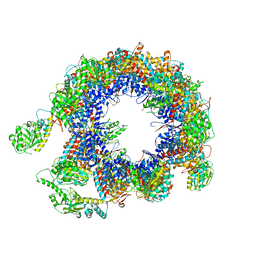 | | TRiC at 0.05 mM ADP-AlFx, Conformation 2, 0.05-C2 | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7NDL
 
 | | Crystal structure of human GFAT-1 S205D | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Isoform 2 of Glutamine-fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
