8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
2PWQ
 
 | | Crystal structure of a putative ubiquitin conjugating enzyme from Plasmodium yoelii | | Descriptor: | Ubiquitin conjugating enzyme | | Authors: | Qiu, W, Dong, A, Hassanali, A, Lin, L, Brokx, S, Altamentova, S, Hills, T, Lew, J, Ravichandran, M, Kozieradzki, I, Zhao, Y, Schapira, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative ubiquitin conjugating enzyme from Plasmodium yoelii.
To be Published
|
|
8B8S
 
 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
6HMB
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6TY5
 
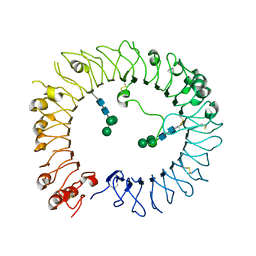 | | Crystal structure of human TLR8 in complex with Compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-7-(7-methyl-2-piperidin-4-yl-indazol-5-yl)furo[3,2-c]pyridin-4-one, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay.
J.Med.Chem., 63, 2020
|
|
5AZZ
 
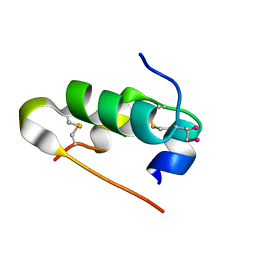 | | Crystal structure of seleno-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Watanabe, S, Okumura, M, Arai, K, Takei, T, Asahina, Y, Hojo, H, Iwaoka, M, Inaba, K. | | Deposit date: | 2015-10-23 | | Release date: | 2017-05-03 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Preparation of Selenoinsulin as a Long-Lasting Insulin Analogue.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5B1D
 
 | |
7KEQ
 
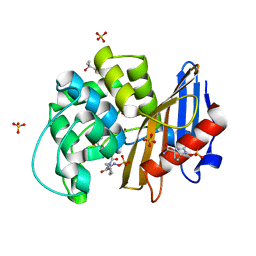 | | avibactam-CDD-1 6 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
2PJ8
 
 | |
1ZWU
 
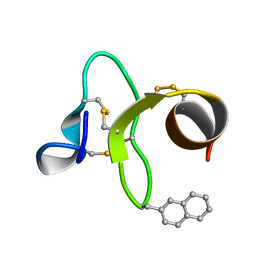 | | 30 NMR structures of AcAMP2-like peptide with non natural beta-(2-naphthyl)-alanine residue. | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 (ACMP2) | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues.
Chemistry, 11, 2005
|
|
1JKT
 
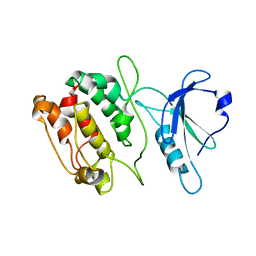 | | TETRAGONAL CRYSTAL FORM OF A CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE | | Descriptor: | DEATH-ASSOCIATED PROTEIN KINASE | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-07-13 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1ATX
 
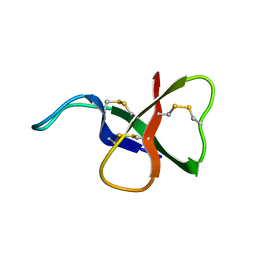 | |
6H03
 
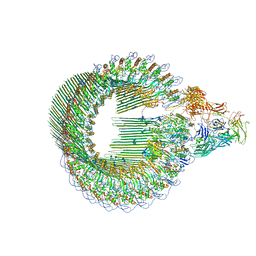 | | OPEN CONFORMATION OF THE MEMBRANE ATTACK COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
1JWF
 
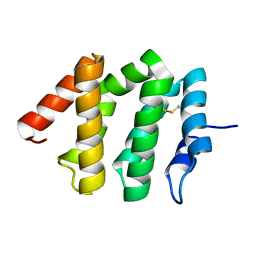 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
6GZA
 
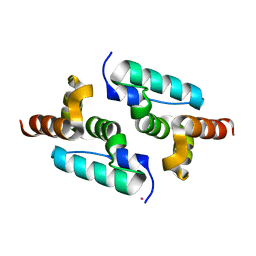 | | Structure of murine leukemia virus capsid C-terminal domain | | Descriptor: | COBALT (II) ION, Capsid protein | | Authors: | Qu, K, Dolezal, M, Schur, F.K.M, Murciano, B, Rumlova, M, Ruml, T, Briggs, J.A.G. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3P4C
 
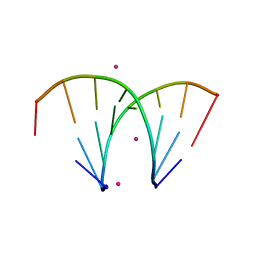 | | Alternatingly modified 2'Fluoro RNA octamer f/rA2U2-R32 | | Descriptor: | 5'-R(*(CFZ)P*GP*(AF2)P*AP*(UFT)P*UP*(CFZ)P*G)-3', STRONTIUM ION | | Authors: | Pallan, P.S, Greene, E.M, Jicman, P.A, Pandey, R.K, Manoharan, M, Rozners, E, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
6H1F
 
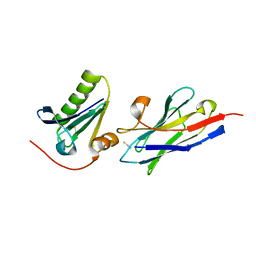 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
6H04
 
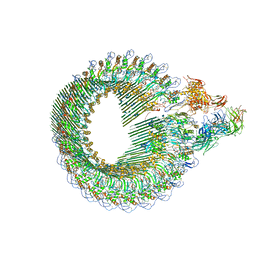 | | Closed conformation of the Membrane Attack Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
4MUA
 
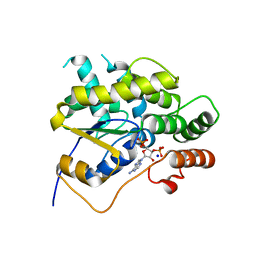 | | Schistosoma mansoni (Blood Fluke) Sulfotransferase | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, Sulfotransferase | | Authors: | Valentim, C.L.L, Cioli, D, Chevalier, F.D, Cao, X, Taylor, A.B, Holloway, S.P, Pica-Mattoccia, L, Guidi, A, Basso, A, Tsai, I.J, Berriman, M, Carvalho-Queiroz, C, Almeida, M, Aguilar, H, Frantz, D.E, Hart, P.J, Anderson, T.J.C, LoVerde, P.T. | | Deposit date: | 2013-09-21 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites.
Science, 342, 2013
|
|
4MVJ
 
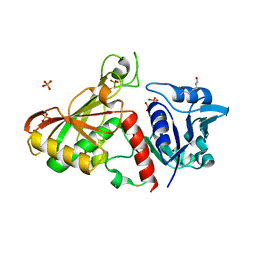 | | 2.85 Angstrom Resolution Crystal Structure of Glyceraldehyde 3-phosphate Dehydrogenase A (gapA) from Escherichia coli Modified by Acetyl Phosphate. | | Descriptor: | ACETATE ION, ACETYLPHOSPHATE, CHLORIDE ION, ... | | Authors: | Minasov, G, Kuhn, M, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
7JIS
 
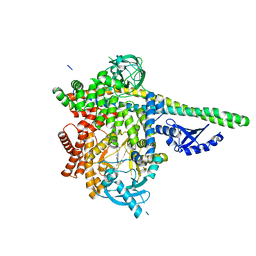 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
1M5Z
 
 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
1LZX
 
 | | Rat neuronal NOS heme domain with NG-hydroxy-L-arginine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
6D8H
 
 | |
