6JVY
 
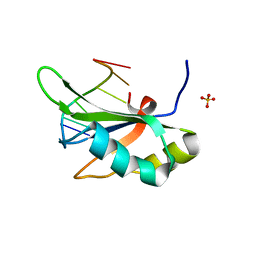 | | Crystal structure of RBM38 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*GP*TP*GP*TP*GP*TP*GP*TP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
5URS
 
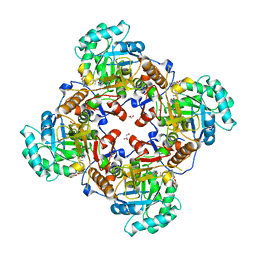 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P178 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P178
To Be Published
|
|
6GJU
 
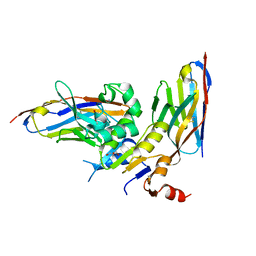 | | human NBD1 of CFTR in complex with nanobodies T2a and T4 | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
5CZB
 
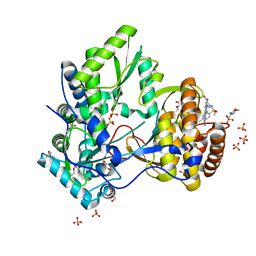 | | HCV NS5B IN COMPLEX WITH LIGAND IDX17119-5 | | Descriptor: | 1-[4-(7-amino-5-methylpyrazolo[1,5-a]pyrimidin-2-yl)phenyl]-3-{[(R)-(2,4-dimethylphenyl)(methoxy)phosphoryl]amino}-1H-pyrazole-4-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pierra, C, Dousson, C, Augustin, M. | | Deposit date: | 2015-07-31 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Synthesis of potent and broad genotypically active NS5B HCV non-nucleoside inhibitors binding to the thumb domain allosteric site 2 of the viral polymerase.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5UUV
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with a product IMP and the inhibitor P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with a product IMP and the inhibitor P182
To Be Published
|
|
5UWX
 
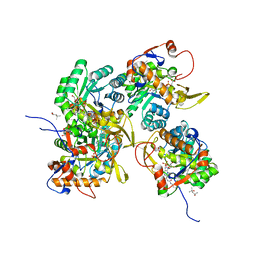 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P176 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P176
To Be Published
|
|
5UXE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P178 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P178
To Be Published
|
|
3N0K
 
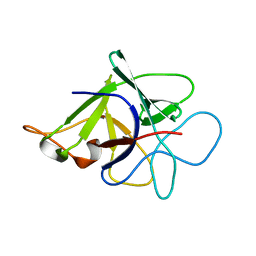 | | Proteinase inhibitor from Coprinopsis cinerea | | Descriptor: | Serine protease inhibitor 1 | | Authors: | Renko, M, Sabotic, J, Bleuler-Martinez, S, Kallert, S, Avanzo, P, Kos, J, Aebi, M, Kuenzler, M, Turk, D. | | Deposit date: | 2010-05-14 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Trypsin Inhibition and Entomotoxicity of Cospin, Serine Protease Inhibitor Involved in Defense of Coprinopsis cinerea Fruiting Bodies.
J.Biol.Chem., 287, 2012
|
|
1Y6Z
 
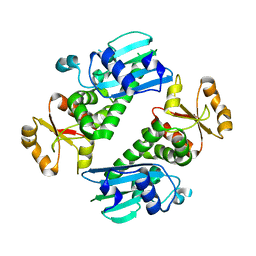 | | Middle domain of Plasmodium falciparum putative heat shock protein PF14_0417 | | Descriptor: | heat shock protein, putative | | Authors: | Lunin, V.V, Botchkareva, E, Loppnau, P, Amani, M, Bray, J, Vedadi, M, Edwards, A, Arrowsmith, C, Sundstrom, M, Bochkarev, A, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-07 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
8BQW
 
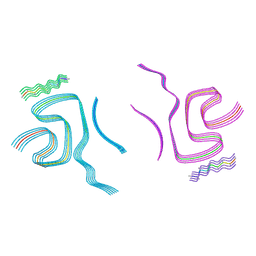 | | Cryo-EM structure of alpha-synuclein filaments doublet from Juvenile-onset synucleinopathy | | Descriptor: | Alpha-synuclein, Unknown protein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
7MJ9
 
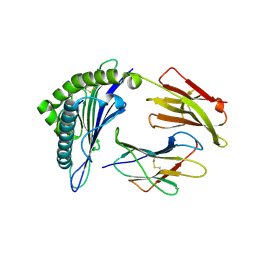 | | HLA-A*02:01 bound to Neuroblastoma Derived mutant IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, Insulin-like growth factor-binding protein-like 1 altered peptide, MHC class I antigen | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
7MJ6
 
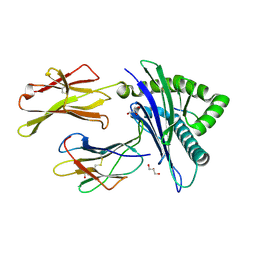 | | HLA-A*02:01 bound to Neuroblastoma Derived IGFBPL1 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Insulin-like growth factor-binding protein-like 1 peptide, ... | | Authors: | Toor, J.S, Tripathi, S.M, Truong, H.V, Yarmarkovich, M, Maris, J.M, Sgourakis, N.G. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Nature, 599, 2021
|
|
8BQV
 
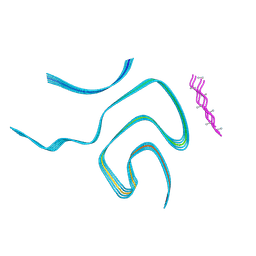 | | Cryo-EM structure of alpha-synuclein singlet filament from Juvenile-onset synucleinopathy | | Descriptor: | Alpha-synuclein, Unknown protein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
1ZOV
 
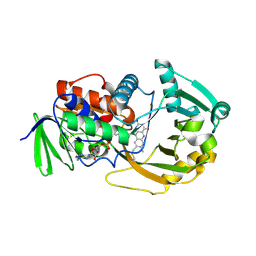 | | Crystal Structure of Monomeric Sarcosine Oxidase from Bacillus sp. NS-129 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Nagata, K, Sasaki, H, Ohtsuka, J, Hua, M, Okai, M, Kubota, K, Kamo, M, Ito, K, Ichikawa, T, Koyama, Y, Tanokura, M. | | Deposit date: | 2005-05-14 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of monomeric sarcosine oxidase from Bacillus sp. NS-129 reveals multiple conformations at the active-site loop
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
6H6O
 
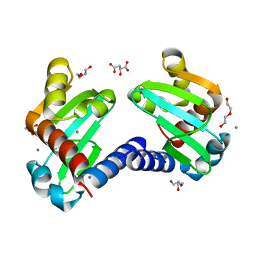 | | UbiJ-SCP2 Ubiquinone synthesis protein | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Legrand, P, Pecqueur, L, Ciccone, L, Lombard, M, Fontecave, M. | | Deposit date: | 2018-07-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone.
Cell Chem Biol, 26, 2019
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
6BYT
 
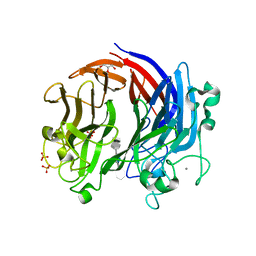 | | Complex structure of LOR107 mutant (R320) with tetrasaccharide substrate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-function analyses of a PL24 family ulvan lyase reveal key features and suggest its catalytic mechanism.
J. Biol. Chem., 293, 2018
|
|
3N9S
 
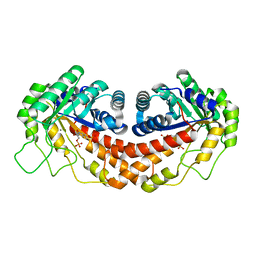 | | Class II fructose-1,6-bisphosphate aldolase from helicobacter pylori in complex with N-(4-hydroxybutyl)- glycolohydroxamic acid bis-phosphate, a competitive inhibitor | | Descriptor: | 4-{hydroxy[(phosphonooxy)acetyl]amino}butyl dihydrogen phosphate, CALCIUM ION, Fructose-bisphosphate aldolase, ... | | Authors: | Coincon, M, Sygusch, S. | | Deposit date: | 2010-05-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Rational Design, Synthesis, and Evaluation of New Selective Inhibitors of Microbial Class II (Zinc Dependent) Fructose Bis-phosphate Aldolases.
J.Med.Chem., 53, 2010
|
|
6GJS
 
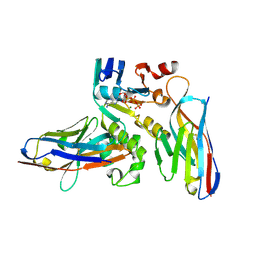 | | Human NBD1 of CFTR in complex with nanobodies D12 and T4 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
6BR4
 
 | | Crystal structure of Escherichia coli DsbA in complex with {N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-2-ylphenyl)methanamine | | Authors: | Heras, B, Totsika, M, Paxman, J.J, Wang, G, Scanlon, M.J, Martin, J.L. | | Deposit date: | 2017-11-29 | | Release date: | 2017-12-27 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibition of Diverse DsbA Enzymes in Multi-DsbA Encoding Pathogens.
Antioxid. Redox Signal., 29, 2018
|
|
6GQQ
 
 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 35) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
6GK4
 
 | | Human NBD1 of CFTR in complex with nanobodies D12 and T8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, GLYCEROL, ... | | Authors: | Sigoillot, M, Overtus, M, Grodecka, M, Scholl, D, Garcia-Pino, A, Laeremans, T, He, L, Pardon, E, Hildebrandt, E, Urbatsch, I, Steyaert, J, Riordan, J.R, Govaerts, C. | | Deposit date: | 2018-05-18 | | Release date: | 2019-06-19 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Domain-interface dynamics of CFTR revealed by stabilizing nanobodies.
Nat Commun, 10, 2019
|
|
3N50
 
 | | Human Early B-cell factor 3 (EBF3) IPT/TIG and HLHLH domains | | Descriptor: | Transcription factor COE3 | | Authors: | Lehtio, L, Siponen, M.I, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schuler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
5GHK
 
 | | Crystal Structure Analysis of Canine serum albumin | | Descriptor: | Serum albumin | | Authors: | Kihira, K, Yamada, K, Kureishi, M, Yokomaku, K, Shinohara, R, Akiyama, M, Komatsu, T. | | Deposit date: | 2016-06-20 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Artificial Blood for Dogs
Sci Rep, 6, 2016
|
|
3NFK
 
 | | Crystal structure of the PTPN4 PDZ domain complexed with the C-terminus of a rabies virus G protein | | Descriptor: | GLYCEROL, Glycoprotein G, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Babault, N, Cordier, F, Lafage, M, Cockburn, J, Haouz, A, Rey, F.A, Delepierre, M, Buc, H, Lafon, M, Wolff, N. | | Deposit date: | 2010-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Peptides Targeting the PDZ Domain of PTPN4 Are Efficient Inducers of Glioblastoma Cell Death.
Structure, 19, 2011
|
|
