8GH7
 
 | | 142D6 bound to BIR3-XIAP | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, BIR3 inhibitor MAA-CHG-PRO-ZHW, E3 ubiquitin-protein ligase XIAP, ... | | Authors: | Garza-Granados, A, McGuire, J, Baggio, C, Pellecchia, M, Pegan, S.D. | | Deposit date: | 2023-03-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a Potent and Orally Bioavailable Lys-Covalent Inhibitor of Apoptosis Protein (IAP) Antagonist.
J.Med.Chem., 66, 2023
|
|
6HM4
 
 | | Crystal structure of Rad4 BRCT1,2 in complex with a Mdb1 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA damage response protein Mdb1, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.770186 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
2R77
 
 | | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum | | Descriptor: | Phosphatidylethanolamine-binding protein, putative | | Authors: | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Crombette, L, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of phosphatidylethanolamine-binding protein, pfl0955c, from Plasmodium falciparum.
To be Published
|
|
6HMC
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 1,2-ETHANEDIOL, 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Dimper, V, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
8GKH
 
 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | Descriptor: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2023-03-19 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
3LMW
 
 | | Crystal structure of iota-carrageenase family GH82 from A. fortis in absence of chloride ions | | Descriptor: | CALCIUM ION, Iota-carrageenase, CgiA, ... | | Authors: | Rebuffet, E, Barbeyron, T, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2010-02-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytic residues and mechanistic analysis of family GH82 iota-carrageenases
Biochemistry, 49, 2010
|
|
5FOG
 
 | | Crystal structure of hte Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of norvaline (Nv2AA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
6T3V
 
 | | Psychrophilic aromatic amino acids aminotransferase from Psychrobacter sp. B6 cocrystalized with substrate analog - malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Rutkiewicz, M, Bujacz, A, Rum, J, Bujacz, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Evidence of Active Site Adaptability towards Different Sized Substrates of Aromatic Amino Acid Aminotransferase from Psychrobacter Sp. B6.
Materials, 14, 2021
|
|
6CWU
 
 | | Protein Tyrosine Phosphatase 1B F135Y mutant | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Hjortness, M, Zwart, P, Sankaran, B, Fox, J.M. | | Deposit date: | 2018-03-31 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Evolutionarily Conserved Allosteric Communication in Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
6T7P
 
 | | human plasmakallikrein protease domain in complex with active site directed inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-6-yl]carbonyl]-~{N}-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Renatus, M. | | Deposit date: | 2019-10-22 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6FDT
 
 | |
6TJO
 
 | | Cryo-EM structure of TypeI tau filaments extracted from the brains of individuals with Corticobasal degeneration | | Descriptor: | Microtubule-associated protein tau | | Authors: | Zhang, W, Murzin, A.G, Falcon, B, Shi, Y, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2019-11-26 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Novel tau filament fold in corticobasal degeneration.
Nature, 580, 2020
|
|
2C0J
 
 | | Crystal structure of the bet3-trs33 heterodimer | | Descriptor: | PALMITIC ACID, R32611_2, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3 | | Authors: | Kim, M.-S, Yi, M.-J, Lee, K.-H, Wagner, J, Munger, C, Kim, Y.-G, Whiteway, M, Cygler, M, Oh, B.-H, Sacher, M. | | Deposit date: | 2005-09-03 | | Release date: | 2006-02-07 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies Reveal a Specific Interaction between Trapp Subunits Trs33P and Bet3P
Traffic, 6, 2005
|
|
6FN3
 
 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
5NCS
 
 | | Structure of the native serpin-type proteinase inhibitor, miropin. | | Descriptor: | Serpin | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
7EUV
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing Flurbiprofen | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, Fatty acid-binding protein, heart | | Authors: | Sugiyama, S, Kakinouchi, K, Hoshina, M, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with Flurbiprofen
To Be Published
|
|
6YRF
 
 | | Vip3Bc1 tetramer | | Descriptor: | Vegetative insecticidal protein | | Authors: | Thompson, R.F, Byrne, M.J, Iadanza, M.I, Arribas Perez, M, Maskell, D.P, George, R.M, Hesketh, E.L, Beales, P.A, Zack, M.D, Berry, C. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane.
Nat Commun, 12, 2021
|
|
2RKF
 
 | | HIV-1 PR resistant mutant + LPV | | Descriptor: | GLYCEROL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PROTEASE RETROPEPSIN, ... | | Authors: | Rezacova, P, Brynda, J, Kozisek, M, Saskova, K.G, Konvalinka, J. | | Deposit date: | 2007-10-16 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ninety-nine is not enough: molecular characterization of inhibitor-resistant human immunodeficiency virus type 1 protease mutants with insertions in the flap region
J.Virol., 82, 2008
|
|
5NJ2
 
 | | Crystal structure of BlaC from Mycobacterium tuberculosis bound to phosphate | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ... | | Authors: | Tassoni, R, Pannu, N.S, Ubbink, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Phosphate Promotes the Recovery of Mycobacterium tuberculosis beta-Lactamase from Clavulanic Acid Inhibition.
Biochemistry, 56, 2017
|
|
1WIB
 
 | | Solution structure of the N-terminal domain from mouse hypothetical protein BAB22488 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain from mouse hypothetical protein BAB22488
To be Published
|
|
7BDQ
 
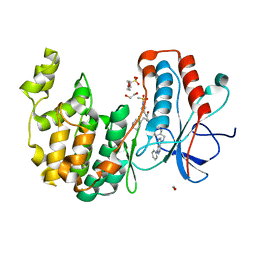 | | MAPK14 bound with SR300 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BE4
 
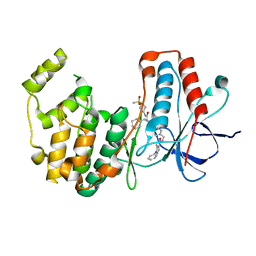 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR159 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7BE5
 
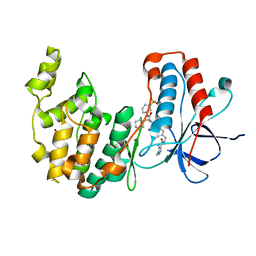 | | Crystal structure of MAP kinase p38 alpha in complex with inhibitor SR276 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{R})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-methyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Joerger, A.C, Schroeder, M, Roehm, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8000524 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
3LVK
 
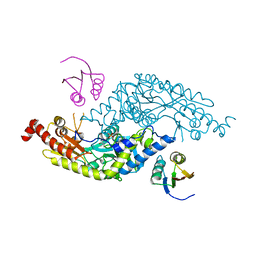 | | Crystal Structure of E.coli IscS-TusA complex (form 2) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
