6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
3P2W
 
 | | Unliganded form of Polo-like kinase I Polo-box domain | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase PLK1 | | Authors: | Sledz, P, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3P34
 
 | | Polo-like kinase I Polo-box domain in complex with MQSpTPL phosphopeptide | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Sledz, P, Hyvonen, M, Abell, C. | | Deposit date: | 2010-10-04 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | From crystal packing to molecular recognition: prediction and discovery of a binding site on the surface of polo-like kinase 1
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
6II1
 
 | | Crystal Structure Analysis of CO form hemoglobin from Bos taurus | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Morita, Y, Yamada, T, Kureishi, M, Komatsu, T. | | Deposit date: | 2018-10-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Quaternary Structure Analysis of a Hemoglobin Core in Hemoglobin-Albumin Cluster.
J Phys Chem B, 122, 2018
|
|
3P45
 
 | | Crystal structure of apo-caspase-6 at physiological pH | | Descriptor: | caspase-6 | | Authors: | Mueller, I, Lamers, M.B.A.C, Ritchie, A.J, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2010-10-06 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures of active and inhibitor-bound human Casp6
To be Published
|
|
6IDK
 
 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class I particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
3P5A
 
 | |
6IEV
 
 | | Crystal structure of a designed protein | | Descriptor: | Designed protein | | Authors: | Han, M, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Protein Sci., 28, 2019
|
|
3OMW
 
 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | Descriptor: | CG14216 | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8701 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
3P6Y
 
 | | CF Im25-CF Im68-UGUAA complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*A)-3', Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
6IJJ
 
 | | Photosystem I of Chlamydomonas reinhardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X, Ma, J, Su, X, Liu, Z, Zhang, X, Li, M. | | Deposit date: | 2018-10-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex.
Nat Plants, 5, 2019
|
|
6IMX
 
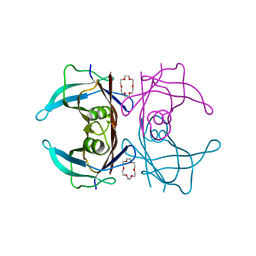 | | Crystal structure of V30M mutated transthyretin in complex with 18-Crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Transthyretin | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitors.
J. Med. Chem., 62, 2019
|
|
6IN4
 
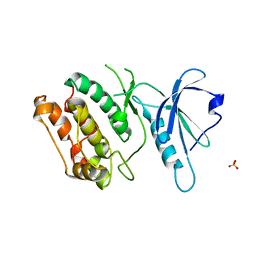 | | Crystal structure of apo DAPK1 in the presence of 18-crown-6 | | Descriptor: | Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
3OK4
 
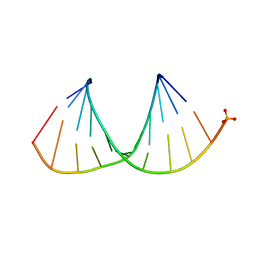 | |
6IJB
 
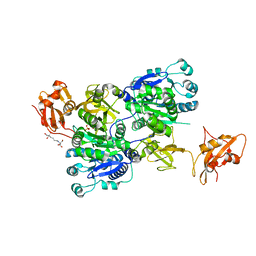 | | Structure of 3-methylmercaptopropionate CoA ligase mutant K523A in complex with AMP and MMPA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(methylsulfanyl)propanoic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
3OSV
 
 | | The crytsal structure of FLGD from P. Aeruginosa | | Descriptor: | Flagellar basal-body rod modification protein FlgD, GLYCEROL | | Authors: | Wang, D, Luo, M, Niu, S. | | Deposit date: | 2010-09-10 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel dimer form of FlgD from P. aeruginosa PAO1
Proteins, 79, 2011
|
|
6IK5
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IR9
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
3OUU
 
 | | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni | | Descriptor: | Biotin carboxylase, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-beta-gamma-ATP Complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
3OWQ
 
 | | X-Ray Structure of Lin1025 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR164 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1025 protein | | Authors: | Kuzin, A, Su, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-20 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR164
To be published
|
|
6INQ
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | Descriptor: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-10-26 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
6I8H
 
 | |
3OZE
 
 | |
3OZD
 
 | | Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase in complex with pCl-phenylthioDADMeImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase
to be published
|
|
