6ANE
 
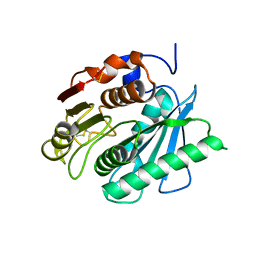 | | Crystal Structure of Ideonella sakaiensis PET Hydrolase | | Descriptor: | MAGNESIUM ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Galaz-Davison, P, Sotomayor, M, Parra, L.P, Ramirez-Sarmiento, C.A. | | Deposit date: | 2017-08-12 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Active Site Flexibility as a Hallmark for Efficient PET Degradation by I. sakaiensis PETase.
Biophys. J., 114, 2018
|
|
1AHW
 
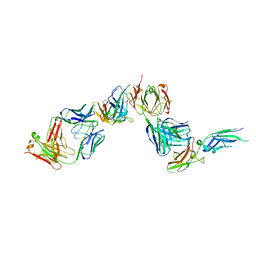 | | A COMPLEX OF EXTRACELLULAR DOMAIN OF TISSUE FACTOR WITH AN INHIBITORY FAB (5G9) | | Descriptor: | IMMUNOGLOBULIN FAB 5G9 (HEAVY CHAIN), IMMUNOGLOBULIN FAB 5G9 (LIGHT CHAIN), TISSUE FACTOR | | Authors: | Huang, M, Syed, R, Stura, E.A, Stone, M.J, Stefanko, R.S, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 1997-04-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mechanism of an inhibitory antibody on TF-initiated blood coagulation revealed by the crystal structures of human tissue factor, Fab 5G9 and TF.5G9 complex.
J.Mol.Biol., 275, 1998
|
|
6ARP
 
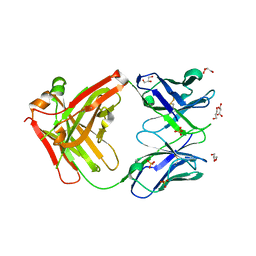 | | Structure of a mutant Cetuximab Fab fragment | | Descriptor: | Cetuximab mutant Fab heavy chain,Immunoglobulin gamma-1 heavy chain, Cetuximab mutant light chain,Uncharacterized protein, GLYCEROL, ... | | Authors: | Christie, M, Christ, D. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a mutant Cetuximab Fab fragment
To Be Published
|
|
1A5T
 
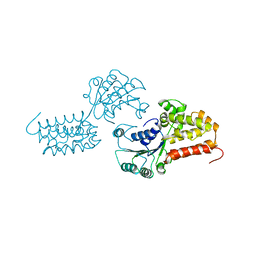 | | CRYSTAL STRUCTURE OF THE DELTA PRIME SUBUNIT OF THE CLAMP-LOADER COMPLEX OF ESCHERICHIA COLI DNA POLYMERASE III | | Descriptor: | DELTA PRIME, ZINC ION | | Authors: | Guenther, B, Onrust, R, Sali, A, O'Donnell, M, Kuriyan, J. | | Deposit date: | 1998-02-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the delta' subunit of the clamp-loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 91, 1997
|
|
1A8E
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Macgillivray, R.T.A, Moore, S.A, Chen, J, Anderson, B.F, Baker, H, Luo, Y, Bewley, M, Smith, C.A, Murphy, M.E.P, Wang, Y, Mason, A.B, Woodworth, R.C, Brayer, G.D, Baker, E.N. | | Deposit date: | 1998-03-24 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
Biochemistry, 37, 1998
|
|
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
6AVJ
 
 | | Crystal structure of human Mitochondrial inner NEET protein (MiNT)/CISD3 | | Descriptor: | CDGSH iron-sulfur domain-containing protein 3, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Lipper, C.H, Karmi, O, Sohn, Y.S, Darash-Yahana, M, Lammert, H, Song, L, Liu, A, Mittler, R, Nechushtai, R, Onuchic, J.N, Jennings, P.A. | | Deposit date: | 2017-09-02 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1AF8
 
 | | ACTINORHODIN POLYKETIDE SYNTHASE ACYL CARRIER PROTEIN FROM STREPTOMYCES COELICOLOR A3(2), NMR, 24 STRUCTURES | | Descriptor: | ACTINORHODIN POLYKETIDE SYNTHASE ACYL CARRIER PROTEIN | | Authors: | Crump, M.P, Crosby, J, Dempsey, C.E, Parkinson, J.A, Murray, M, Hopwood, D.A, Simpson, T.J. | | Deposit date: | 1997-03-23 | | Release date: | 1997-09-26 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2).
Biochemistry, 36, 1997
|
|
6AWE
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2-fluoro-6-(2-pyridin-2-ylethylamino)benzonitrile | | Descriptor: | 2-fluoro-6-{[2-(pyridin-2-yl)ethyl]amino}benzonitrile, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2-fluoro-6-(2-pyridin-2-ylethylamino)benzonitrile
To Be Published
|
|
1AAR
 
 | | STRUCTURE OF A DIUBIQUITIN CONJUGATE AND A MODEL FOR INTERACTION WITH UBIQUITIN CONJUGATING ENZYME (E2) | | Descriptor: | DI-UBIQUITIN | | Authors: | Cook, W.J, Jeffrey, L.C, Carson, M, Chen, Z, Pickart, C.M. | | Deposit date: | 1992-04-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2).
J.Biol.Chem., 267, 1992
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1AY1
 
 | | ANTI TAQ FAB TP7 | | Descriptor: | TP7 FAB | | Authors: | Murali, R, Helmer-Citterich, M, Sharkey, D.J, Scalice, E.R, Daiss, J.L, Sullivan, M.A, Murthy, H.M.K. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on an inhibitory antibody against Thermus aquaticus DNA polymerase suggest mode of inhibition.
Protein Eng., 11, 1998
|
|
1B0S
 
 | | BINDING OF AR-1-144, A TRI-IMIDAZOLE DNA MINOR GROOVE BINDER, TO CCGG SEQUENCE ANALYZED BY NMR SPECTROSCOPY | | Descriptor: | (2-{[4-({4-[(4-FORMYLAMINO-1-METHYL-1H-IMIDAZOLE-2-CARBONYL)-AMINO]-1-METHYL-1H-IMIDAZOLE-2-CARBONYL}-AMINO)-1-METHYL-1 H-IMIDAZOLE-2-CARBONYL]-AMINO}-ETHYL)-DIMETHYL-AMMONIUM, DNA (5'-D(*GP*AP*AP*CP*CP*GP*GP*TP*TP*C)-3') | | Authors: | Yang, X.-L, Kaenzig, C, Lee, M, Wang, A.H.-J. | | Deposit date: | 1998-11-12 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Binding of AR-1-144, a tri-imidazole DNA minor groove binder, to CCGG sequence analyzed by NMR spectroscopy.
Eur.J.Biochem., 263, 1999
|
|
1B90
 
 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1B12
 
 | | CRYSTAL STRUCTURE OF TYPE 1 SIGNAL PEPTIDASE FROM ESCHERICHIA COLI IN COMPLEX WITH A BETA-LACTAM INHIBITOR | | Descriptor: | PHOSPHATE ION, SIGNAL PEPTIDASE I, prop-2-en-1-yl (2S)-2-[(2S,3R)-3-(acetyloxy)-1-oxobutan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylate | | Authors: | Paetzel, M, Dalbey, R, Strynadka, N.C.J. | | Deposit date: | 1999-11-24 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor.
Nature, 396, 1998
|
|
7UYX
 
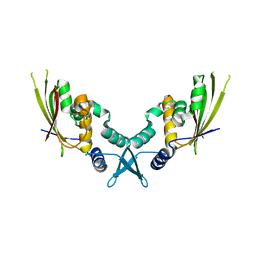 | | Structure of bacteriophage PA1c gp2 | | Descriptor: | Bacteriophage PA1C gp2 | | Authors: | Enustun, E, Deep, A, Gu, Y, Nguyen, K, Chaikeeratisak, V, Armbruster, E, Ghassemian, M, Pogliano, J, Corbett, K.D. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of the bacteriophage nucleus protein interaction network.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1B9O
 
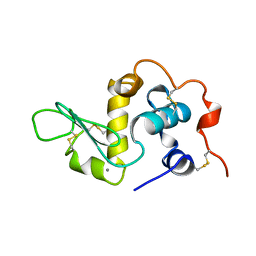 | | HUMAN ALPHA-LACTALBUMIN, LOW TEMPERATURE FORM | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Harata, K, Abe, Y, Muraki, M. | | Deposit date: | 1999-02-14 | | Release date: | 1999-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic evaluation of internal motion of human alpha-lactalbumin refined by full-matrix least-squares method.
J.Mol.Biol., 287, 1999
|
|
1B9V
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN TEH ACTIVE SITE | | Descriptor: | 1-[4-CARBOXY-2-(3-PENTYLAMINO)PHENYL]-5,5'-DI(HYDROXYMETHYL)PYRROLIDIN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zahao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1AUG
 
 | | CRYSTAL STRUCTURE OF THE PYROGLUTAMYL PEPTIDASE I FROM BACILLUS AMYLOLIQUEFACIENS | | Descriptor: | PYROGLUTAMYL PEPTIDASE-1 | | Authors: | Odagaki, Y, Hayashi, A, Okada, K, Hirotsu, K, Kabashima, T, Ito, K, Yoshimoto, T, Tsuru, D, Sato, M, Clardy, J. | | Deposit date: | 1997-08-26 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyroglutamyl peptidase I from Bacillus amyloliquefaciens reveals a new structure for a cysteine protease.
Structure Fold.Des., 7, 1999
|
|
1AZV
 
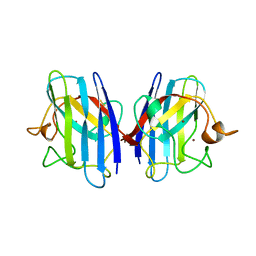 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
1B9S
 
 | | NOVEL AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE MAKE SELECTIVE INTERACTIONS WITH CONSERVED RESIDUES AND WATER MOLECULES IN THE ACTIVE SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-ACETYLAMINO)-3-[N-(2-ETHYLBUTANOYLAMINO)]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Finley, J.B, Atigadda, V.R, Duarte, F, Zhao, J.J, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 1999-02-15 | | Release date: | 1999-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel aromatic inhibitors of influenza virus neuraminidase make selective interactions with conserved residues and water molecules in the active site.
J.Mol.Biol., 293, 1999
|
|
1AOE
 
 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 1,3-DIAMINO-7-(1-ETHYEPROPYE)-7H-PYRRALO-[3,2-F]QUINAZOLINE (GW345) | | Descriptor: | 7-(1-ETHYL-PROPYL)-7H-PYRROLO-[3,2-F]QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
1B6X
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE GUANINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 4 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3' | | Authors: | Cullinan, D, Johnson, F, Grollman, A.P, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyguanosine.
Biochemistry, 36, 1997
|
|
1BA4
 
 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
1BHZ
 
 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF HEN EGG WHITE LYSOZYME FROM MASC DATA | | Descriptor: | LYSOZYME | | Authors: | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
