1F6B
 
 | | CRYSTAL STRUCTURE OF SAR1-GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SAR1, ... | | Authors: | Huang, M, Wilson, I.A, Balch, W.E. | | Deposit date: | 2000-06-21 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export.
J.Cell Biol., 155, 2001
|
|
8P9O
 
 | | PCNA from Chaetomium thermophilum in complex with PolD3 peptide | | Descriptor: | Proliferating cell nuclear antigen, Synthetic peptide corresponding to amino acids 437 to 451 of PolD3 from Chaetomium thermophilum | | Authors: | Alphey, M.S, Wolford, C.B, MacNeill, S.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Canonical binding of Chaetomium thermophilum DNA polymerase delta / zeta subunit PolD3 and flap endonuclease Fen1 to PCNA.
Front Mol Biosci, 10, 2023
|
|
7UUG
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7UUP
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8JH2
 
 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | Descriptor: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
8JH3
 
 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH4
 
 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8RHJ
 
 | | Yeast 20S proteasome in complex with a macrocyclic oxindole epoxyketone (compound 5) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Macrocyclic oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
1B8L
 
 | | Calcium-bound D51A/E101D/F102W Triple Mutant of Beta Carp Parvalbumin | | Descriptor: | CALCIUM ION, CARBONATE ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-01 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
8RGZ
 
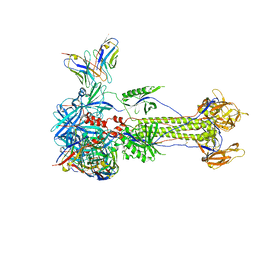 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH0
 
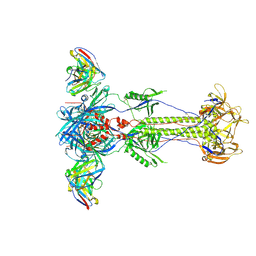 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8S8A
 
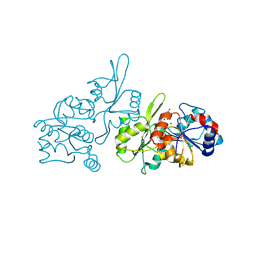 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone without phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CHLORIDE ION, Chronophin, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
8RPX
 
 | | NhoI restriction endonuclease in complex with quadruply methylated DNA target | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*TP*GP*(5CM)P*AP*GP*(5CM)P*TP*C)-3'), ... | | Authors: | Rafalski, D, Krakowska, K, Gilski, M, Bochtler, M. | | Deposit date: | 2024-01-17 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural analysis of the BisI family of modification dependent restriction endonucleases.
Nucleic Acids Res., 52, 2024
|
|
8OY0
 
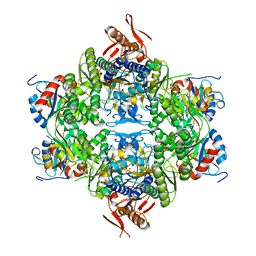 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Acinetobacter baumanii | | Descriptor: | ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, GLYCEROL, ... | | Authors: | Alphey, M.S, Read, B, da Silva, R.G. | | Deposit date: | 2023-05-03 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.
Biochemistry, 63, 2024
|
|
1FNM
 
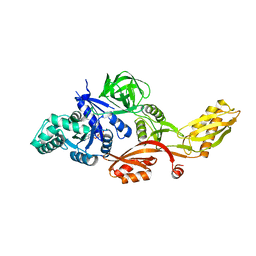 | | STRUCTURE OF THERMUS THERMOPHILUS EF-G H573A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Laurberg, M, Kristensen, O, Martemyanov, K, Gudkov, A.T, Nagaev, I, Hughes, D, Liljas, A. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site.
J.Mol.Biol., 303, 2000
|
|
1C0L
 
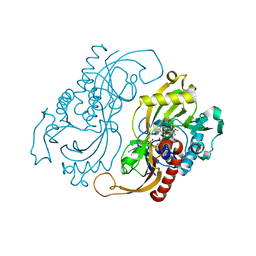 | | D-AMINO ACID OXIDASE: STRUCTURE OF SUBSTRATE COMPLEXES AT VERY HIGH RESOLUTION REVEAL THE CHEMICAL REACTTION MECHANISM OF FLAVIN DEHYDROGENATION | | Descriptor: | D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, TRIFLUOROALANINE | | Authors: | Umhau, S, Molla, G, Diederichs, K, Pilone, M.S, Ghisla, S, Welte, W. | | Deposit date: | 1999-07-16 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C3P
 
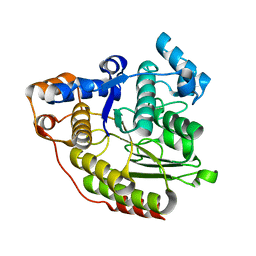 | |
8P5L
 
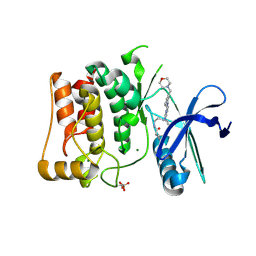 | | Kinase domain of mutant human ULK1 in complex with compound MRT67307 | | Descriptor: | MAGNESIUM ION, N-{3-[(5-cyclopropyl-2-{[3-(morpholin-4-ylmethyl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase ULK1 | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8P5H
 
 | | Kinase domain of mutant human ULK1 in complex with compound CCT241533 | | Descriptor: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8J4C
 
 | | YeeE(TsuA)-YeeD(TsuB) complex for thiosulfate uptake | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Spirochaeta thermophila YeeE(TsuA)-YeeD(TsuB),UPF0033 domain-containing protein, SirA-like domain-containing protein (chimera), ... | | Authors: | Ikei, M, Miyazaki, R, Monden, K, Naito, Y, Takeuchi, A, Takahashi, Y.S, Tanaka, Y, Ichikawa, M, Tsukazaki, T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | YeeD is an essential partner for YeeE-mediated thiosulfate uptake in bacteria and regulates thiosulfate ion decomposition.
Plos Biol., 22, 2024
|
|
8P5G
 
 | | Kinase domain of wild type human ULK1 in complex with compound CCT241533 | | Descriptor: | 4-FLUORO-2-(4-{[(3S,4R)-4-(1-HYDROXY-1-METHYLETHYL)PYRROLIDIN-3-YL]AMINO}-6,7-DIMETHOXYQUINAZOLIN-2-YL)PHENOL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8P5I
 
 | | Kinase domain of mutant human ULK1 in complex with compound XMD-17-51 | | Descriptor: | 5,11-dimethyl-2-[(1-piperidin-4-ylpyrazol-4-yl)amino]pyrimido[4,5-b][1,4]benzodiazepin-6-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.829 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8P5K
 
 | | Kinase domain of mutant human ULK1 in complex with compound MRT68921 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
8P5J
 
 | | Kinase domain of mutant human ULK1 in complex with compound WZ4003 | | Descriptor: | MAGNESIUM ION, SODIUM ION, Serine/threonine-protein kinase ULK1, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
