6B6F
 
 | |
9B0E
 
 | | Cryo-EM Structure of E.coli produced recombinant N-acetyltransferase 10 (NAT10) in complex with cytidine-amide-CoA bisubstrate probe and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA cytidine acetyltransferase, [[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{R})-4-[[3-[2-[2-[[1-[(2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-2-oxidanylidene-pyrimidin-4-yl]amino]-2-oxidanylidene-ethyl]sulfanylethylamino]-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Zhou, M, Marmorstein, R. | | Deposit date: | 2024-03-12 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Molecular Basis for RNA Cytidine Acetylation by NAT10.
Biorxiv, 2024
|
|
9GNM
 
 | |
9GNL
 
 | | X-ray crystal structure of VvPYL1-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Rivera-Moreno, M, Benavente, J.L, Infantes, L, Albert, A. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of transpiration in grapevine through chemical activation of ABA signaling by ABA receptor agonists
To Be Published
|
|
5OI2
 
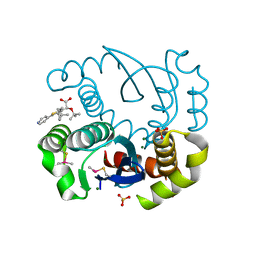 | |
8ZDW
 
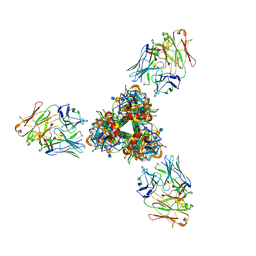 | | The cryoEM structure of H5N1 HA split from symmetric filament in conformation A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain, ... | | Authors: | Li, R, Gao, J, Wang, L, Gui, M, Xiang, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Multivalent interactions between fully glycosylated influenza virus hemagglutinins mediated by glycans at distinct N-glycosylation sites
To Be Published
|
|
5OI3
 
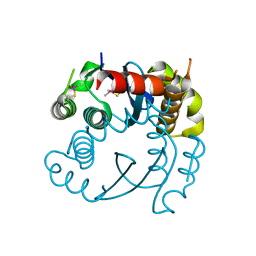 | |
9FCG
 
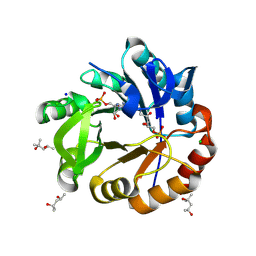 | | Medicago truncatula 5'-ProFAR isomerase (HISN3) D57N mutant in complex with PrFAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic, ... | | Authors: | Witek, W, Imiolczyk, B, Ruszkowski, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural, kinetic, and evolutionary peculiarities of HISN3, a plant 5'-ProFAR isomerase.
Plant Physiol Biochem., 215, 2024
|
|
9CER
 
 | | Guillardia theta Fanzor (GtFz) State 1 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Guillardia theta Fanzor1, RNA (142-MER), ZINC ION | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9CF2
 
 | | Parasitella parasitica Fanzor (PpFz) State 3 | | Descriptor: | DNA non-target strand, DNA substrate model, DNA target strand, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
5OIA
 
 | |
9FGO
 
 | | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site | | Descriptor: | CHLORIDE ION, Polyprotein, ZINC ION | | Authors: | Ni, X, Koekemoer, L, Williams, E.P, Wang, S, Wright, N.D, Godoy, A.S, Aschenbrenner, J.C, Balcomb, B.H, Lithgo, R.M, Marples, P.G, Fairhead, M, Thompson, W, Kirkegaard, K, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-05-24 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of Enterovirus 71 2A protease mutant C110A containing VP1-2A junction in the active site
To Be Published
|
|
9FCY
 
 | | CysG(N-16)-R21K mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | ADENINE, CALCIUM ION, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
5OI9
 
 | | Trichoplax adhaerens STIL N-terminal domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
9CP7
 
 | |
5OIX
 
 | | Structure of the HMPV P oligomerization domain at 1.6 A | | Descriptor: | GLYCEROL, Phosphoprotein | | Authors: | Renner, M, Paesen, G.C, Grison, C.M, Granier, S, Grimes, J.M, Leyrat, C. | | Deposit date: | 2017-07-19 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural dissection of human metapneumovirus phosphoprotein using small angle x-ray scattering.
Sci Rep, 7, 2017
|
|
9EMU
 
 | | RosC-8-demethyl-8-amino-FMN - Phosphate complex | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Ermler, U, Mack, M, Demmer, U. | | Deposit date: | 2024-03-11 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Phosphatase RosC from Streptomyces davaonensis is Used for Roseoflavin Biosynthesis and has Evolved to Largely Prevent Dephosphorylation of the Important Cofactor Riboflavin-5'-phosphate.
J.Mol.Biol., 436, 2024
|
|
9EQJ
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-39 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[cis-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9BKD
 
 | | The structure of human Pdcd4 bound to the 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Albacete-Albacete, L, Ramakrishnan, V, S.Fraser, C. | | Deposit date: | 2024-04-27 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
9FCU
 
 | | CysG(N-16)-H121A mutant in complex with SAH from Kitasatospora cystarginea | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the SAM- and metal-dependent mechanism of O-methyltransferases in cystargolide and belactosin biosynthesis: A structure-activity relationship study.
J.Biol.Chem., 300, 2024
|
|
8XX8
 
 | | Structure of Glycilhalorhodopsin from Salinarimonas soli | | Descriptor: | CHLORIDE ION, EICOSANE, OLEIC ACID, ... | | Authors: | Suzuki, K, Ishizuka, T, Konno, M, Inoue, K, Murata, T. | | Deposit date: | 2024-01-17 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Light-driven anion-pumping rhodopsin with unique cytoplasmic anion-release mechanism.
J.Biol.Chem., 300, 2024
|
|
9J4L
 
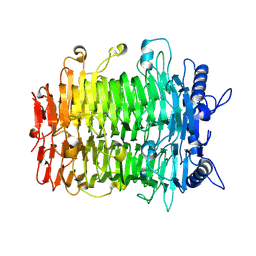 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | Descriptor: | DFA-III-forming inulin fructotransferase | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9FCF
 
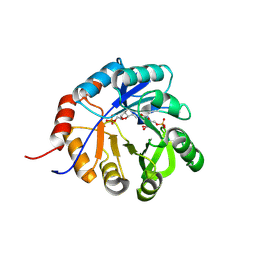 | | Medicago truncatula 5'-ProFAR isomerase (HISN3) D57N mutant in complex with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic, CHLORIDE ION, ... | | Authors: | Witek, W, Imiolczyk, B, Ruszkowski, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural, kinetic, and evolutionary peculiarities of HISN3, a plant 5'-ProFAR isomerase.
Plant Physiol Biochem., 215, 2024
|
|
8GFT
 
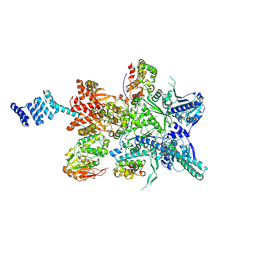 | | Hsp90 provides platform for CRaf dephosphorylation by PP5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, ... | | Authors: | Jaime-Garza, M, Nowotny, C.A, Coutandin, D, Wang, F, Tabios, M, Agard, D.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Hsp90 provides a platform for kinase dephosphorylation by PP5.
Nat Commun, 14, 2023
|
|
9IWQ
 
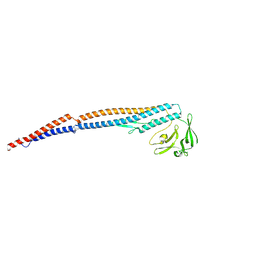 | | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament | | Descriptor: | Flagellin | | Authors: | Waraich, K, Makino, F, Miyata, T, Kinoshita, M, Minamino, T, Namba, K. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament
To Be Published
|
|
