4IH8
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.877 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
3GCJ
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid
To be Published
|
|
4MQM
 
 | | Crystal structure of Antigen 85C in presence of Ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
3GD1
 
 | | Structure of an Arrestin/Clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking | | Descriptor: | Beta-arrestin-1, Clathrin heavy chain 1, clathrin | | Authors: | Kang, D.S, Kern, R.C, Puthenveedu, M.A, von Zastrow, M, Williams, J.C, Benovic, J.L. | | Deposit date: | 2009-02-23 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an arrestin2-clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking
J.Biol.Chem., 284, 2009
|
|
4MR9
 
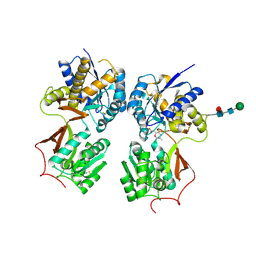 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist SCH50911 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-17 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
4IJ0
 
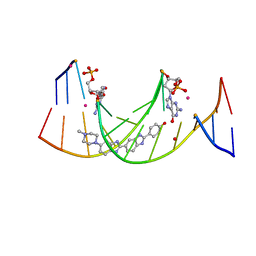 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
1KP6
 
 | | USTILAGO MAYDIS KILLER TOXIN KP6 ALPHA-SUBUNIT | | Descriptor: | PROTEIN (TOXIN), SULFATE ION | | Authors: | Li, N, Erman, M, Pangborn, W, Duax, W.L, Park, C.-M, Bruenn, J, Ghosh, D. | | Deposit date: | 1999-05-28 | | Release date: | 1999-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ustilago maydis killer toxin KP6 alpha-subunit. A multimeric assembly with a central pore.
J.Biol.Chem., 274, 1999
|
|
1KOR
 
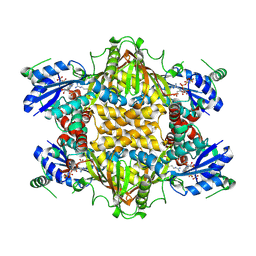 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitors | | Descriptor: | ARGININE, Argininosuccinate Synthetase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Goto, M, Nakajima, Y, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of argininosuccinate synthetase from Thermus thermophilus HB8. Structural basis for the catalytic action.
J.Biol.Chem., 277, 2002
|
|
4MS4
 
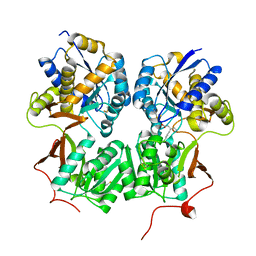 | | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the agonist baclofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Geng, Y, Bush, M, Mosyak, L, Wang, F, Fan, Q.R. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural mechanism of ligand activation in human GABA(B) receptor.
Nature, 504, 2013
|
|
3S9Q
 
 | | Crystal structure of native type 1 ribosome inactivating protein from Momordica balsamina at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
3SE7
 
 | | ancient VanA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, VanA | | Authors: | Wright, G.D, Morar, M. | | Deposit date: | 2011-06-10 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Antibiotic resistance is ancient.
Nature, 477, 2011
|
|
3SFD
 
 | | crystal structure of porcine mitochondrial respiratory complex II bound with oxaloacetate and pentachlorophenol | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhou, Q.J, Zhai, Y.J, Liu, M, Sun, F. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Protein Cell, 2, 2011
|
|
3SDF
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipoteichoic acid at 2.1 A Resolution | | Descriptor: | (2S)-1-({3-O-[2-(acetylamino)-4-amino-2,4,6-trideoxy-beta-D-galactopyranosyl]-alpha-D-glucopyranosyl}oxy)-3-(heptanoyloxy)propan-2-yl (7Z)-pentadec-7-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Lipoteichoic acid at 2.1 A Resolution
To be Published
|
|
1KQ8
 
 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
5RF8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z271004858 | | Descriptor: | 3C-like proteinase, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102179 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[3-(2-oxopyrrolidin-1-yl)phenyl]acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4IB8
 
 | | Bovine beta-lactoglobulin (isoform A) in complex with dodecyl sulphate (SDS) | | Descriptor: | DODECYL SULFATE, beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Swiatek, S, Dziedzicka-Wasylewska, M, Lewinski, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The differences in binding 12-carbon aliphatic ligands by bovine beta-lactoglobulin isoform A and B studied by isothermal titration calorimetry and X-ray crystallography
J.Mol.Recognit., 26, 2013
|
|
5REH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z111507846 | | Descriptor: | 1-cyclohexyl-3-(2-pyridin-4-ylethyl)urea, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3SES
 
 | | Cu-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
4MVE
 
 | |
5REX
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102287 | | Descriptor: | 1-{4-[(naphthalen-1-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RF6
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1348371854 | | Descriptor: | 3C-like proteinase, 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1KW9
 
 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 2.0A resolution | | Descriptor: | 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-29 | | Release date: | 2003-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
4IBX
 
 | | Crystal structure of stabilized TEM-1 beta-lactamase variant v.13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, CALCIUM ION, ... | | Authors: | Dellus-Gur, E, Toth-Petroczy, A, Elias, M, Tawfik, D.S. | | Deposit date: | 2012-12-09 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | What Makes a Protein Fold Amenable to Functional Innovation? Fold Polarity and Stability Trade-offs.
J.Mol.Biol., 425, 2013
|
|
1KSK
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
