4N2I
 
 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 10 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
2O95
 
 | | Crystal Structure of the Metal-Free Dimeric Human Mov34 MPN domain (residues 1-186) | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 26S proteasome non-ATPase regulatory subunit 7, DODECAETHYLENE GLYCOL, ... | | Authors: | Sanches, M, Alves, B.S.C, Zanchin, N.I.T, Guimaraes, B.G. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Mov34 MPN Domain Reveals a Metal-free Dimer
J.Mol.Biol., 370, 2007
|
|
3MOL
 
 | | Structure of dimeric holo HasAp H32A Mutant from Pseudomonas aeruginosa to 1.20A Resolution | | Descriptor: | Heme acquisition protein HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Jepkorir, G, Rodriguez, J.C, Rui, H, Im, W, Alontaga, A.Y, Yukl, E, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2010-04-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, NMR Spectroscopic, and Computational Investigation of Hemin Loading in the Hemophore HasAp from Pseudomonas aeruginosa.
J.Am.Chem.Soc., 132, 2010
|
|
3MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH UNMODIFIED DNA AND ADOHCY | | Descriptor: | DNA (5'-D(*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | Authors: | O'Gara, M, Klimasauskas, S, Roberts, R.J, Cheng, X. | | Deposit date: | 1996-07-24 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
1LU4
 
 | | 1.1 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A SECRETED MYCOBACTERIUM TUBERCULOSIS DISULFIDE OXIDOREDUCTASE HOMOLOGOUS TO E. COLI DSBE: IMPLICATIONS FOR FUNCTIONS | | Descriptor: | SOLUBLE SECRETED ANTIGEN MPT53 | | Authors: | Goulding, C.W, Apostol, M.I, Gleiter, S, Parseghian, A, Bardwell, J, Gennaro, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-21 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Gram-positive DsbE Proteins Function Differently from Gram-negative DsbE Homologs: A STRUCTURE TO FUNCTION ANALYSIS OF DsbE FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 279, 2004
|
|
5C8I
 
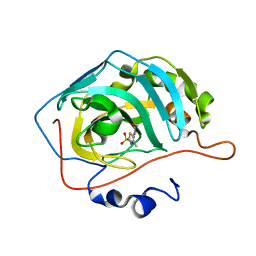 | | Joint X-ray/neutron structure of Human Carbonic Anhydrase II in complex with Methazolamide | | Descriptor: | Carbonic anhydrase 2, N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2(3H)-ylidene)acetamide, ZINC ION | | Authors: | Aggarwal, M, Kovalevsky, A.Y, Fisher, S.Z, McKenna, R. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.56 Å), X-RAY DIFFRACTION | | Cite: | Neutron structure of human carbonic anhydrase II in complex with methazolamide: mapping the solvent and hydrogen-bonding patterns of an effective clinical drug.
IUCrJ, 3, 2016
|
|
3MHZ
 
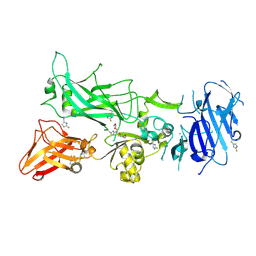 | | 1.7A structure of 2-fluorohistidine labeled Protective Antigen | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Wimalasena, D.S, Janowiak, B.E, Miyagi, M, Sun, J, Hajduch, J, Pooput, C, Kirk, K.L, Bann, J.G. | | Deposit date: | 2010-04-09 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence that histidine protonation of receptor-bound anthrax protective antigen is a trigger for pore formation.
Biochemistry, 49, 2010
|
|
1L3G
 
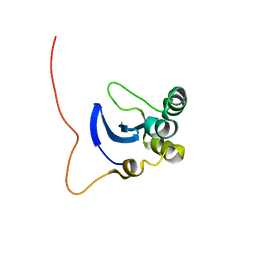 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | Descriptor: | TRANSCRIPTION FACTOR Mbp1 | | Authors: | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
6FI6
 
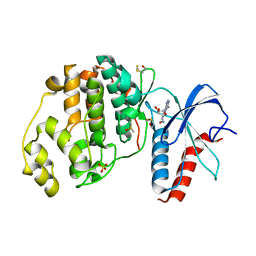 | | Crystal structure of ERK2 in complex with an adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-azanyl-8-diazanyl-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2018-01-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of ERK2 in complex with an adenosine derivative
To be published
|
|
8A6M
 
 | |
3MKU
 
 | | Structure of a Cation-bound Multidrug and Toxin Compound Extrusion (MATE) transporter | | Descriptor: | Multi antimicrobial extrusion protein (Na(+)/drug antiporter) MATE-like MDR efflux pump, RUBIDIUM ION | | Authors: | He, X, Szewczyk, P, Karyakin, A, Evin, M, Hong, W.-X, Zhang, Q, Chang, G. | | Deposit date: | 2010-04-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of a cation-bound multidrug and toxic compound extrusion transporter.
Nature, 467, 2010
|
|
1KGN
 
 | | R2F from Corynebacterium Ammoniagenes in its oxidised, Fe containing, form | | Descriptor: | FE (III) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
3MSE
 
 | | Crystal structure of C-terminal domain of PF110239. | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, putative, ... | | Authors: | Wernimont, A.K, Artz, J.D, Hutchinson, A, Sullivan, H, Weadge, J, Tempel, W, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Lin, Y.H, Neculai, A.M, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of C-terminal domain of PF110239
To be Published
|
|
1KHR
 
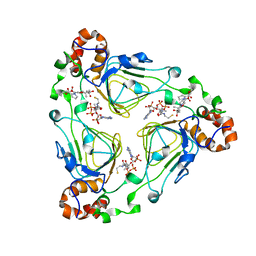 | |
6FJ3
 
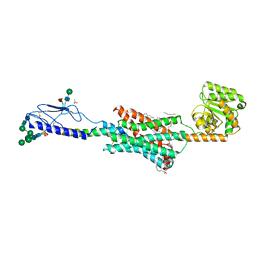 | | High resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist. | | Descriptor: | ACETIC ACID, CHLORIDE ION, OLEIC ACID, ... | | Authors: | Ehrenmann, J, Schoppe, J, Klenk, C, Rappas, M, Kummer, L, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High-resolution crystal structure of parathyroid hormone 1 receptor in complex with a peptide agonist.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6FLD
 
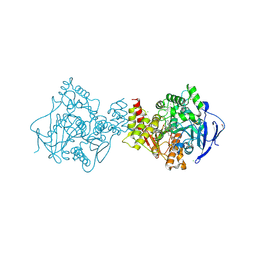 | | Carbamylated T. californica acetylcholineterase bound to uncharged hybrid reactivator 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, ... | | Authors: | De la Mora, E, Santoni, G, de Souza, J, Sussman, J, Silman, I, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-01-25 | | Release date: | 2018-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
6HOK
 
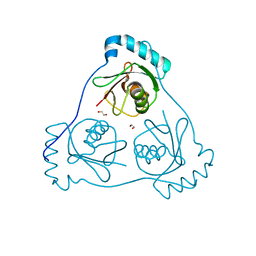 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
5CJV
 
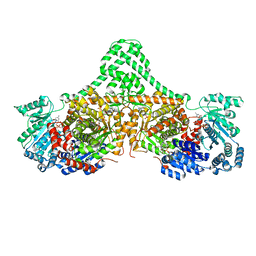 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate isovaleryl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
6HWH
 
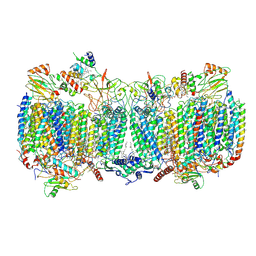 | | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | | Descriptor: | CARDIOLIPIN, COPPER (II) ION, Co-purified unknown peptide built as polyALA, ... | | Authors: | Wiseman, B, Nitharwal, R.G, Fedotovskaya, O, Schafer, J, Guo, H, Kuang, Q, Benlekbir, S, Sjostrand, D, Adelroth, P, Rubinstein, J.L, Brzezinski, P, Hogbom, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-11-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4MQ3
 
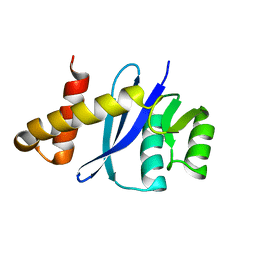 | |
5CM2
 
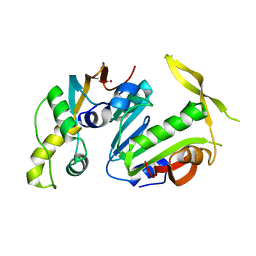 | | Structure of Y. lipolytica Trm9-Trm112 complex, a methyltransferase modifying U34 in the anticodon loop of some tRNAs | | Descriptor: | TRNA METHYLTRANSFERASE, TRNA METHYLTRANSFERASE ACTIVATOR SUBUNIT, ZINC ION | | Authors: | Letoquart, J, van Tran, N, Caroline, V, Aleksandrov, A, Lazar, N, Van Tilbeurgh, H, Liger, D, Graille, M. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure.
Nucleic Acids Res., 43, 2015
|
|
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
6HZ3
 
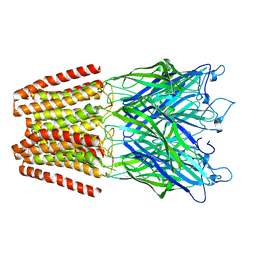 | | THE GLIC PENTAMERIC LIGAND-GATED ION CHANNEL MUTANT E243G | | Descriptor: | CHLORIDE ION, Proton-gated ion channel | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Electrostatics, proton sensor, and networks governing the gating transition in GLIC, a proton-gated pentameric ion channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G88
 
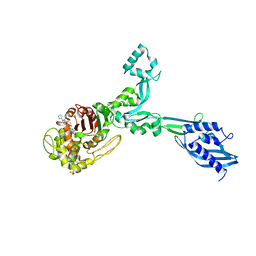 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
5CJT
 
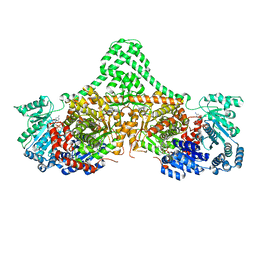 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate isobutyryl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
