4LFG
 
 | | Crystal structure of geranylgeranyl diphosphate synthase sub1274 (target efi-509455) from streptococcus uberis 0140j with bound magnesium and isopentyl diphosphate, fully liganded complex; | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl Diphosphate Synthase, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Geranylgeranyl Diphosphate Synthase from Streptococcus Uberis 0140J
To be Published
|
|
6F9L
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | Authors: | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
6FAN
 
 | |
1LMM
 
 | | Solution Structure of Psmalmotoxin 1, the First Characterized Specific Blocker of ASIC1a NA+ channel | | Descriptor: | Psalmotoxin 1 | | Authors: | Escoubas, P, Bernard, C, Lazdunski, M, Darbon, H. | | Deposit date: | 2002-05-02 | | Release date: | 2003-11-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Recombinant production and solution structure of PcTx1, the specific peptide inhibitor of ASIC1a proton-gated cation channels
Protein Sci., 12, 2003
|
|
6H8J
 
 | | 1.45 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPTO | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-08-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into Urease Inhibition by N-( n-Butyl) Phosphoric Triamide through an Integrated Structural and Kinetic Approach.
J.Agric.Food Chem., 67, 2019
|
|
1LFV
 
 | | OXY HEMOGLOBIN (88% RELATIVE HUMIDITY) | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Biswal, B.K, Vijayan, M. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human oxy- and deoxyhaemoglobin at different levels of humidity: variability in the T state.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5A0W
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI E117A IN COMPLEX WITH ITS TARGET DNA AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, ACETATE ION, CHLORIDE ION, ... | | Authors: | Molina, R, Besker, N, Prieto, J, Montoya, G, D'Abramo, M. | | Deposit date: | 2015-04-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Dynamical Characterization of the I- Dmo Catalytic Activity
To be Published
|
|
1LO6
 
 | | Human Kallikrein 6 (hK6) active form with benzamidine inhibitor at 1.56 A resolution | | Descriptor: | BENZAMIDINE, Kallikrein 6, MAGNESIUM ION | | Authors: | Bernett, M.J, Blaber, S.I, Scarisbrick, I.A, Dhanarajan, P, Thompson, S.M, Blaber, M. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure and biochemical characterization of human kallikrein 6 reveals a
trypsin-like kallikrein is expressed in the central nervous system
To be Published
|
|
3IB7
 
 | | Crystal structure of full length Rv0805 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A mycobacterial cyclic AMP phosphodiesterase that moonlights as a modifier of cell wall permeability
J.Biol.Chem., 284, 2009
|
|
6JKA
 
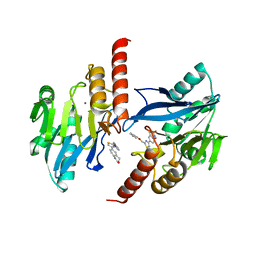 | | Crystal structure of metallo-beta-lactamse, IMP-1, in complex with a thiazole-bearing inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-(phenoxymethyl)-1,3-thiazol-4-yl]phenyl]propanoic acid, 6-[2-(phenoxymethyl)-1,3-thiazol-4-yl]-3,4-dihydro-1H-quinolin-2-one, Metallo-beta-lactamase type 2, ... | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
4YST
 
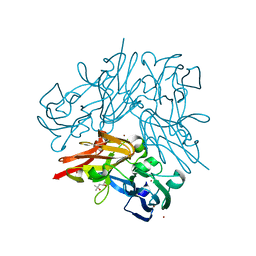 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
6JKQ
 
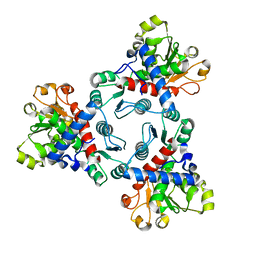 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi (Ligand-free form) | | Descriptor: | Aspartate carbamoyltransferase | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
4LSX
 
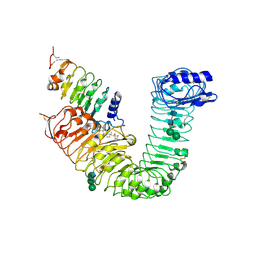 | | Plant steroid receptor ectodomain bound to brassinolide and SERK1 co-receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, ... | | Authors: | Santiago, J, Henzler, C, Hothorn, M. | | Deposit date: | 2013-07-23 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases.
Science, 341, 2013
|
|
1LQ7
 
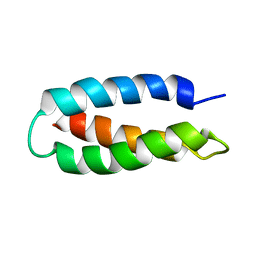 | | De Novo Designed Protein Model of Radical Enzymes | | Descriptor: | Alpha3W | | Authors: | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | Deposit date: | 2002-05-09 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
3MIU
 
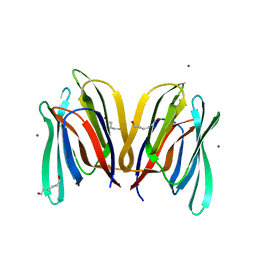 | | Structure of Banana Lectin-pentamannose complex | | Descriptor: | HEXANE-1,6-DIOL, Lectin, ZINC ION, ... | | Authors: | Sharma, A, Vijayan, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Influence of glycosidic linkage on the nature of carbohydrate binding in beta-prism I fold lectins: an X-ray and molecular dynamics investigation on banana lectin-carbohydrate complexes
Glycobiology, 21, 2011
|
|
3MIV
 
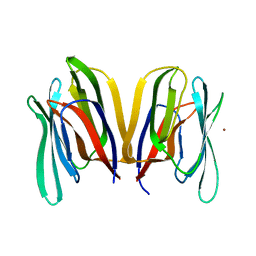 | | Structure of Banana lectin - Glc-alpha(1,2)-Glc complex | | Descriptor: | Lectin, ZINC ION | | Authors: | Sharma, A, Vijayan, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Influence of glycosidic linkage on the nature of carbohydrate binding in beta-prism I fold lectins: an X-ray and molecular dynamics investigation on banana lectin-carbohydrate complexes
Glycobiology, 21, 2011
|
|
6JL6
 
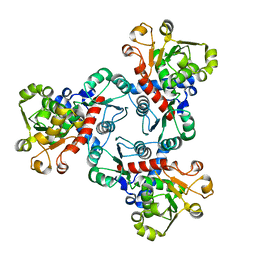 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, PHOSPHATE ION | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6EZG
 
 | | Torpedo californica AChE in complex with indolic multi-target directed ligand | | Descriptor: | 1-(7-chloranyl-4-methoxy-1~{H}-indol-5-yl)-3-[1-(phenylmethyl)piperidin-4-yl]propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, Lalut, J, Karila, D, Lecoutey, C, Davis, A, Nachon, F, Sillman, I, Sussman, J, Weik, M, Maurice, T, Dallemagne, P, Rochais, C. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel multitarget-directed ligands targeting acetylcholinesterase and sigma1receptors as lead compounds for treatment of Alzheimer's disease: Synthesis, evaluation, and structural characterization of their complexes with acetylcholinesterase.
Eur J Med Chem, 162, 2018
|
|
7ZGO
 
 | | Cryo-EM structure of human NKCC1 (TM domain) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nissen, P, Fenton, R, Neumann, C, Lindtoft Rosenbaek, L, Kock Flygaard, R, Habeck, M, Lykkegaard Karlsen, J, Wang, Y, Lindorff-Larsen, K, Gad, H, Hartmann, R, Lyons, J. | | Deposit date: | 2022-04-04 | | Release date: | 2022-10-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the human NKCC1 transporter reveals mechanisms of ion coupling and specificity.
Embo J., 41, 2022
|
|
6EZS
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylglucosaminidase Nag2, MALONATE ION | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
5A4J
 
 | | Crystal structure of FTHFS1 from T.acetoxydans Re1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, D(-)-TARTARIC ACID, ... | | Authors: | Bergdahl, R, Jacobson, F, Muller, B, Mikkelsen, N, Schurer, A, Sandgren, M. | | Deposit date: | 2015-06-10 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Characterization, Crystallization and Three- Dimensional Structures of Formyltetrahydrofolate Synthetase (Fthfs) from the Syntrophic Acetate Oxidising Bacterium Tepidanaerobacter Acetatoxydans Re1
To be Published
|
|
5A6W
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with the HMA domain of Pikp1 from rice (Oryza sativa) | | Descriptor: | 1,2-ETHANEDIOL, AVR-PIK PROTEIN, RESISTANCE PROTEIN PIKP-1, ... | | Authors: | Maqbool, A, Saitoh, H, Franceschetti, M, Stevenson, C.E, Uemura, A, Kanzaki, H, Kamoun, S, Terauchi, R, Banfield, M.J. | | Deposit date: | 2015-07-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor.
Elife, 4, 2015
|
|
1KYT
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
6F0J
 
 | | GLIC mutant E26A | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Hu, H.D, Delarue, M. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Full mutational mapping of titratable residues helps to identify proton-sensors involved in the control of channel gating in the Gloeobacter violaceus pentameric ligand-gated ion channel.
PLoS Biol., 15, 2017
|
|
5A72
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | Descriptor: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
