3ZG3
 
 | | STEROL 14-ALPHA DEMETHYLASE (CYP51)FROM TRYPANOSOMA CRUZI IN COMPLEX WITH THE PYRIDINE INHIBITOR N-(1-(5-(trifluoromethyl)(pyridin-2-yl)) piperidin-4yl)-N-(4-(trifluoromethyl)phenyl)pyridin-3-amine (EPL- BS967, UDD) | | Descriptor: | N-[4-(trifluoromethyl)phenyl]-N-[1-[5-(trifluoromethyl)pyridin-2-yl]piperidin-4-yl]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Keenan, M, Chatelain, E, Lepesheva, G.I. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complexes of Trypanosoma Cruzi Sterol 14Alpha-Demethylase (Cyp51) with Two Pyridine-Based Drug Candidates for Chagas Disease: Structural Basis for Pathogen-Selectivity
J.Biol.Chem., 288, 2013
|
|
3ZHQ
 
 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD | | Descriptor: | CALCIUM ION, MAGNESIUM ION, MULTIFUNCTIONAL 2-OXOGLUTARATE METABOLISM ENZYME, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
6U87
 
 | | Pseudomonas aeruginosa HasA mutant - Y75H | | Descriptor: | HasAp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Brimberry, M, Lanzilotta, W, Wilks, A, Dent, A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Axial Heme Coordination by the Tyr-His Motif in the Extracellular Hemophore HasAp Is Critical for the Release of Heme to the HasR Receptor of Pseudomonas aeruginosa .
Biochemistry, 60, 2021
|
|
3ZWN
 
 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZXG
 
 | | lysenin sphingomyelin complex | | Descriptor: | LYSENIN, SULFATE ION, TRIMETHYL-[2-[[(2S,3S)-2-(OCTADECANOYLAMINO)-3-OXIDANYL-BUTOXY]-OXIDANYL-PHOSPHORYL]OXYETHYL]AZANIUM | | Authors: | De Colibus, L, Sonnen, A.F.P, Morris, K.J, Siebert, C.A, Abrusci, P, Plitzko, J, Hodnik, V, Leippe, M, Volpi, E, Anderluh, G, Gilbert, R.J.C. | | Deposit date: | 2011-08-10 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of Lysenin Reveal a Shared Evolutionary Origin for Pore-Forming Proteins and its Mode of Sphingomyelin Recognition.
Structure, 20, 2012
|
|
3ZXT
 
 | |
6U8Q
 
 | | CryoEM structure of HIV-1 cleaved synaptic complex (CSC) intasome | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-09-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
3ZZH
 
 | | Crystal structure of the amino acid kinase domain from Saccharomyces cerevisiae acetylglutamate kinase in complex with its feed- back inhibitor L-arginine | | Descriptor: | 1,2-ETHANEDIOL, ACETYLGLUTAMATE KINASE, ARGININE, ... | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
3ZV8
 
 | |
6UE0
 
 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
3ZVF
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 85 | | Descriptor: | 3C PROTEASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVS
 
 | |
3ZWV
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase.
J. Mol. Biol., 415, 2012
|
|
7RL8
 
 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
3ZXY
 
 | | Structure of S218A mutant of the protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
6TV0
 
 | | Serratia spp. cyanase hydratase | | Descriptor: | Cyanate hydratase, GLYCEROL, OXALIC ACID | | Authors: | Pederzoli, R, Tarantino, D, Gourlay, L.J, Chaves-Sanjuan, A, Bolognesi, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detecting the nature and solving the crystal structure of a contaminant protein from an opportunistic pathogen.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TUU
 
 | | Leishmania infantum Rad51 surrogate LiRadA10 in complex with 5,6,7,8-tetrahydro-2-naphthoic acid | | Descriptor: | 5,6,7,8-tetrahydronaphthalene-2-carboxylic acid, CHLORIDE ION, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of dedicated crystallographic systems for structure-guided drug discovery
To Be Published
|
|
3ZZP
 
 | | Circular permutant of ribosomal protein S6, lacking edge strand beta- 2 of wild-type S6. | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Saraboji, K, Haglund, E, Lindberg, M.O, Oliveberg, M, Logan, D.T. | | Deposit date: | 2011-09-02 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Trimming Down a Protein Structure to its Bare Foldons: Spatial Organization of the Cooperative Unit.
J.Biol.Chem., 287, 2012
|
|
3ZV9
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZWP
 
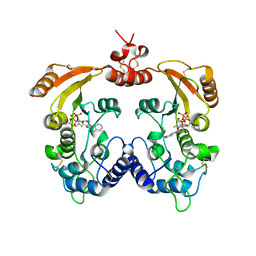 | | Crystal structure of ADP ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.1 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, GLYCEROL, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZHV
 
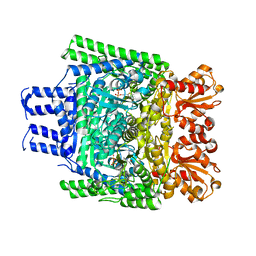 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, post-decarboxylation intermediate from pyruvate (2-hydroxyethyl-ThDP) | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2-[(1S)-1-oxidanylethyl]-1,3-thiazol-3-ium-5-yl]ethyl phosphono hydrogen phosphate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
3ZOW
 
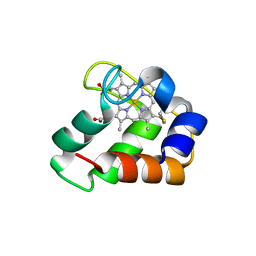 | | Crystal Structure of Wild Type Nitrosomonas europaea Cytochrome c552 | | Descriptor: | CYTOCHROME C-552, HEME C | | Authors: | Hersleth, H.-P, Can, M, Krucinska, J, Zoppellaro, G, Andersen, N.H, Karlsen, S, Wedekind, J.E, Andersson, K.K, Bren, K.L. | | Deposit date: | 2013-02-25 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Nitrosomonas Europaea Cytochrome C-552 Variants with Marked Differences in Electronic Structure.
Chembiochem, 14, 2013
|
|
3ZVA
 
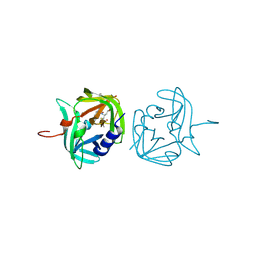 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
6U2Z
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, 1,2-ETHANEDIOL, COPPER (II) ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-21 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the hydrolyzed moxalactam and two copper ions
To Be Published
|
|
