3ATQ
 
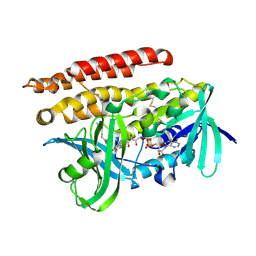 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, TETRADECANE | | Authors: | Sasaki, D, Fujihashi, M, Murakami, M, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
8P8M
 
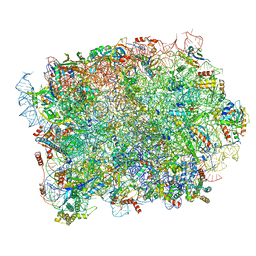 | | Yeast 60S ribosomal subunit, RPL39 deletion | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Yeast 60S ribosomal subunit, RPL39 deletion
To Be Published
|
|
8P9B
 
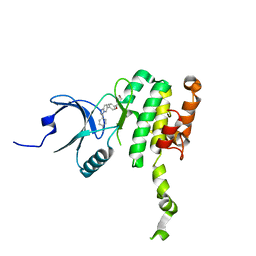 | | Crystal Structure of Mnk2-D228G in complex with Tinodasertib | | Descriptor: | 4-[6-(4-morpholin-4-ylcarbonylphenyl)imidazo[1,2-a]pyridin-3-yl]benzenecarbonitrile, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Turnbull, A.P, Sabin, V, Bell, C, Watson, M. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of Mnk2-D228G in complex with Tinodasertib
To Be Published
|
|
5HTV
 
 | |
3APO
 
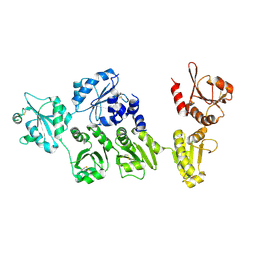 | | Crystal structure of full-length ERdj5 | | Descriptor: | DnaJ homolog subfamily C member 10 | | Authors: | Inaba, K, Suzuki, M, Nagata, K. | | Deposit date: | 2010-10-20 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5.
Mol.Cell, 41, 2011
|
|
6YU5
 
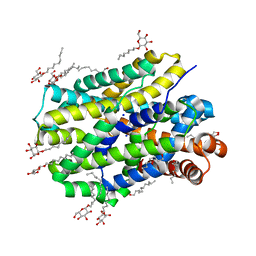 | | Crystal structure of MhsT in complex with L-valine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, SODIUM ION, Sodium-dependent transporter, ... | | Authors: | Focht, D, Neumann, C, Lyons, J, Eguskiza Bilbao, A, Blunck, R, Malinauskaite, L, Schwarz, I.O, Javitch, J.A, Quick, M, Nissen, P. | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A non-helical region in transmembrane helix 6 of hydrophobic amino acid transporter MhsT mediates substrate recognition.
Embo J., 40, 2021
|
|
5KQW
 
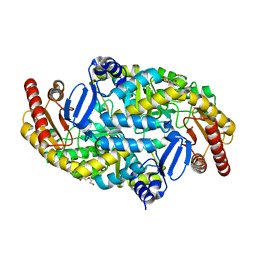 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KND
 
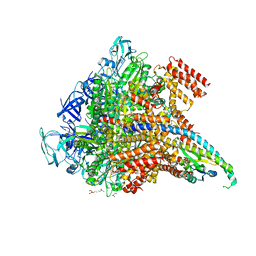 | | Crystal structure of the Pi-bound V1 complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Suzuki, K, Mizutani, K, Maruyama, S, Shimono, K, Imai, F.L, Muneyuki, E, Kakinuma, Y, Ishizuka-Katsura, Y, Shirouzu, M, Yokoyama, S, Yamato, I, Murata, T. | | Deposit date: | 2016-06-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Crystal structures of the ATP-binding and ADP-release dwells of the V1 rotary motor
Nat Commun, 7, 2016
|
|
7LBE
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | Descriptor: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
3UU6
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-22' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ATP
 
 | | Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr with ligand | | Descriptor: | Methyl-accepting chemotaxis protein I, SERINE | | Authors: | Tajima, H, Sakuma, M, Homma, K, Kawagishi, I, Imada, K. | | Deposit date: | 2011-01-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 286, 2011
|
|
7LBG
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
5HV9
 
 | | Human LTC4S mutant-S36E | | Descriptor: | GLUTATHIONE, Leukotriene C4 synthase, SULFATE ION | | Authors: | Thulasingam, M, Ahmad, H.R.S, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2016-01-28 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphorylation of Leukotriene C4 Synthase at Serine 36 Impairs Catalytic Activity.
J.Biol.Chem., 291, 2016
|
|
6YVT
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with MD-253 | | Descriptor: | 2-[[5-(6-methoxynaphthalen-2-yl)-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Egl nine homolog 1, GLYCEROL, ... | | Authors: | Chowdhury, R, Demetriades, M, Schofield, C.J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Dynamic combinatorial chemistry employing boronic acids/boronate esters leads to potent oxygenase inhibitors.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7LBF
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-01-07 | | Release date: | 2021-03-10 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
4I38
 
 | | Structures of IT intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I3I
 
 | | Structures of IT intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected at 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
5KUH
 
 | | GluK2EM with LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Meyerson, J.R, Chittori, S, Merk, A, Rao, P, Han, T.H, Serpe, M, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
8P8U
 
 | | Yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Yeast 60S ribosomal subunit
To Be Published
|
|
3FLW
 
 | | P38 kinase crystal structure in complex with pamapimod | | Descriptor: | 6-(2,4-difluorophenoxy)-2-{[3-hydroxy-1-(2-hydroxyethyl)propyl]amino}-8-methylpyrido[2,3-d]pyrimidin-7(8H)-one, Mitogen-activated protein kinase 14 | | Authors: | Kuglstatter, A, Ghate, M. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Discovery of Pamapimod, R1503 and R1487 as Orally Bioavailable and Highly Selective Inhibitors of p38 Map Kinase
To be Published
|
|
5JQC
 
 | | Crystal structure putative autolysin from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1076 protein, ... | | Authors: | Chang, C, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal structure putative autolysin from Listeria monocytogenes
To Be Published
|
|
4NGJ
 
 | | Dialyzed HEW lysozyme batch crystallized in 1.0 M RbCl and collected at 100 K | | Descriptor: | CHLORIDE ION, Lysozyme C, RUBIDIUM ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3FMJ
 
 | |
5HN6
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and 3-isopropylmalate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-ISOPROPYLMALIC ACID, Homoisocitrate dehydrogenase, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
