5PB0
 
 | | Crystal Structure of Factor VIIa in complex with 2-(4-ethoxy-3-methoxyphenyl)-2-(isoquinolin-6-ylamino)acetic acid | | 分子名称: | (2~{R})-2-(4-ethoxy-3-methoxy-phenyl)-2-(isoquinolin-6-ylamino)ethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | 登録日 | 2016-11-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal Structure of a Factor VIIa complex
To be published
|
|
3D1P
 
 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | 分子名称: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | 著者 | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-06 | | 公開日 | 2008-07-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
7W33
 
 | | The crystal structure of human CtsL in complex with 14a | | 分子名称: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | 著者 | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | 登録日 | 2021-11-25 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
3CZ2
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera at pH 7.0 | | 分子名称: | CHLORIDE ION, Pheromone-binding protein ASP1 | | 著者 | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | 登録日 | 2008-04-27 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | 分子名称: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | 著者 | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | 登録日 | 2021-11-25 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
7W7O
 
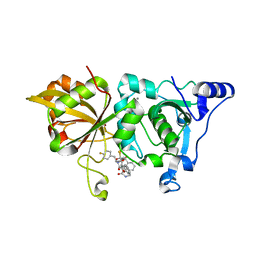 | | The crystal structure of human Calpain-1 protease core in complex with 14a | | 分子名称: | CALCIUM ION, Calpain-1 catalytic subunit, HYDROSULFURIC ACID, ... | | 著者 | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | 登録日 | 2021-12-06 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The crystal structure of human Calpain-1 protease core in complex with 14a
To Be Published
|
|
3CZA
 
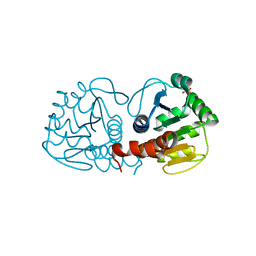 | | Crystal Structure of E18D DJ-1 | | 分子名称: | MALONIC ACID, Protein DJ-1 | | 著者 | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hashim, S, Pozharski, E, Wilson, M.A. | | 登録日 | 2008-04-28 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
3D2Q
 
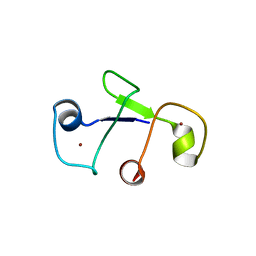 | |
3D1E
 
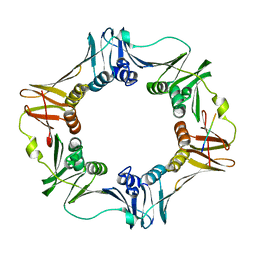 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | 分子名称: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | 著者 | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | 登録日 | 2008-05-05 | | 公開日 | 2008-07-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D2R
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 4 in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kato, M, Wynn, R.M, Chuang, L.C, Tso, S.-C, Li, J, Chuang, D.T. | | 登録日 | 2008-05-08 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Pyruvate Dehydrogenase Kinase-4 Structures Reveal a Metastable Open Conformation Fostering Robust Core-free Basal Activity.
J.Biol.Chem., 283, 2008
|
|
3D5N
 
 | | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125. | | 分子名称: | Q97W15_SULSO | | 著者 | Vorobiev, S.M, Chen, Y, Seetharaman, J, Lee, D, Foote, R.E, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-05-16 | | 公開日 | 2008-07-15 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125.
To be Published
|
|
5PAB
 
 | | Crystal Structure of Factor VIIa in complex with 1-[[3-[2-hydroxy-3-(1H-pyrrolo[3,2-c]pyridin-2-yl)phenyl]phenyl]methyl]-3-phenylurea | | 分子名称: | 1-[[3-[2-oxidanyl-3-(1~{H}-pyrrolo[3,2-c]pyridin-2-yl)phenyl]phenyl]methyl]-3-phenyl-urea, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | 登録日 | 2016-11-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal Structure of a Factor VIIa complex
To be published
|
|
5PAR
 
 | | Crystal Structure of Factor VIIa in complex with 1H-benzimidazol-2-amine | | 分子名称: | 1H-benzimidazol-2-amine, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | 登録日 | 2016-11-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of a Factor VIIa complex
To be published
|
|
5OYC
 
 | | GH5 endo-xyloglucanase from Cellvibrio japonicus | | 分子名称: | CHLORIDE ION, Cellulase, putative, ... | | 著者 | Attia, M, Nelson, C.E, Offen, W.A, Jain, N, Gardner, J.G, Davies, G.J, Brumer, H. | | 登録日 | 2017-09-08 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | In vitro and in vivo characterization of threeCellvibrio japonicusglycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions.
Biotechnol Biofuels, 11, 2018
|
|
7X78
 
 | | L-fuculose 1-phosphate aldolase | | 分子名称: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | 著者 | Lou, X, Zhang, Q, Bartlam, M. | | 登録日 | 2022-03-09 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
7WZE
 
 | |
7X87
 
 | | The complex structure of beta-1,2-glucosyltransferase from Ignavibacterium album with sophotetraose observed as sophorose | | 分子名称: | Beta-galactosidase, CALCIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | 著者 | Kobayashi, K, Shimizu, H, Tanaka, N, Kuramochi, K, Nakai, H, Nakajima, M, Taguchi, H. | | 登録日 | 2022-03-11 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Characterization and structural analyses of a novel glycosyltransferase acting on the beta-1,2-glucosidic linkages.
J Biol Chem, 298, 2022
|
|
3D45
 
 | | Crystal structure of mouse PARN in complex with m7GpppG | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Poly(A)-specific ribonuclease PARN | | 著者 | Wu, M, Song, H. | | 登録日 | 2008-05-13 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease.
Structure, 17, 2009
|
|
7WU5
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | 分子名称: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7XIU
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-14 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
3CER
 
 | | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13 | | 分子名称: | Possible exopolyphosphatase-like protein, SULFATE ION | | 著者 | Kuzin, A.P, Su, M, Chen, Y, Neely, H, Seetharaman, J, Shastry, R, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-02-29 | | 公開日 | 2008-04-01 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the exopolyphosphatase-like protein Q8G5J2. Northeast Structural Genomics Consortium target BlR13.
To be Published
|
|
7XIT
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-14 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ7
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-15 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ5
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | 分子名称: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | 著者 | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | 登録日 | 2022-04-15 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7WU3
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | 分子名称: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
