4TQX
 
 | | Molecular Basis of Streptococcus mutans Sortase A Inhibition by Chalcone. | | 分子名称: | ACETIC ACID, SULFATE ION, Sortase, ... | | 著者 | Wallock-Richards, D.J, Marles-Wright, J, Clarke, D.J, Maitra, A, Dodds, M, Hanley, B, Campopiano, D.J. | | 登録日 | 2014-06-12 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone.
Chem.Commun.(Camb.), 51, 2015
|
|
4OE9
 
 | |
4RWE
 
 | | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92 | | 分子名称: | CHLORIDE ION, GLYCEROL, Sugar-binding transport protein | | 著者 | Tan, K, Zhou, M, Clancy, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-12-03 | | 公開日 | 2014-12-31 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The crystal structure of a sugar-binding transport protein from Yersinia pestis CO92
To be Published
|
|
4OEK
 
 | | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A Resolution | | 分子名称: | 1,2-ETHANEDIOL, 2-PHENYLETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kumar, M, Singh, R.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2014-01-13 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A
To be Published
|
|
4RX1
 
 | | Crystal Structure of antibiotic-resistance methyltransferase Kmr | | 分子名称: | GLYCEROL, IODIDE ION, Putative rRNA methyltransferase | | 著者 | Savic, M. | | 登録日 | 2014-12-08 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.472 Å) | | 主引用文献 | 30S Subunit-Dependent Activation of the Sorangium cellulosum So ce56 Aminoglycoside Resistance-Conferring 16S rRNA Methyltransferase Kmr.
Antimicrob.Agents Chemother., 59, 2015
|
|
4TT6
 
 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | 分子名称: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | 著者 | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | 登録日 | 2014-06-19 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | 分子名称: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | 著者 | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | 登録日 | 2014-06-20 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4RZD
 
 | | Crystal Structure of a PreQ1 Riboswitch | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | 著者 | Wedekind, J.E, Liberman, J.A, Salim, M. | | 登録日 | 2014-12-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RNO
 
 | |
4TOD
 
 | | 2.05A resolution structure of BfrB (D34F) from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | 登録日 | 2014-06-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TPR
 
 | | Structure of Tau5 antibody Fab fragment | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, CHLORIDE ION, ... | | 著者 | Cehlar, O, Skrabana, R, Novak, M. | | 登録日 | 2014-06-09 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Tau5 antibody Fab fragment
To Be Published
|
|
4TSX
 
 | |
4RNY
 
 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | 分子名称: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | 著者 | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | 登録日 | 2014-10-27 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4TPG
 
 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | 分子名称: | Ala-L-3-Br-Tyr-Ala, DODECYL-BETA-D-MALTOSIDE, Proton:oligopeptide symporter POT family, ... | | 著者 | Guettou, F, Quistgaard, E.M, Raba, M, Moberg, P, Low, C, Nordlund, P. | | 登録日 | 2014-06-07 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.91 Å) | | 主引用文献 | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6QDI
 
 | | anti-sigma factor domain-containing protein from Clostridium clariflavum | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, PA14 domain-containing protein | | 著者 | Voronov, M, Bayer, E.A, Livnah, O. | | 登録日 | 2019-01-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Distinctive ligand-binding specificities of tandem PA14 biomass-sensory elements from Clostridium thermocellum and Clostridium clariflavum.
Proteins, 87, 2019
|
|
4RTA
 
 | |
4N9M
 
 | | Joint neutron/x-ray structure of urate oxidase in complex with 8-hydroxyxanthine | | 分子名称: | 8-hydroxy-3,9-dihydro-1H-purine-2,6-dione, CHLORIDE ION, SODIUM ION, ... | | 著者 | Oksanen, E, Blakeley, M.P, Budayova-Spano, M. | | 登録日 | 2013-10-21 | | 公開日 | 2014-02-05 | | 最終更新日 | 2018-06-13 | | 実験手法 | NEUTRON DIFFRACTION (2.023 Å), X-RAY DIFFRACTION | | 主引用文献 | The neutron structure of urate oxidase resolves a long-standing mechanistic conundrum and reveals unexpected changes in protonation.
Plos One, 9, 2014
|
|
4TTV
 
 | | Crystal structure of human ThrRS complexing with a bioengineered macrolide BC194 | | 分子名称: | (1R,2R)-2-[(2S,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadec-6-en-2-yl]cyclobutanecarboxylic acid, Threonine--tRNA ligase, cytoplasmic, ... | | 著者 | Fang, P, Guo, M. | | 登録日 | 2014-06-23 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Aminoacyl-tRNA synthetase dependent angiogenesis revealed by a bioengineered macrolide inhibitor.
Sci Rep, 5, 2015
|
|
4TOA
 
 | | 1.95A resolution structure of Iron Bound BfrB (N148L) from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | 登録日 | 2014-06-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4RUC
 
 | | Crystal structure of Y-family DNA polymerase Dpo4 extending from a MeFapy-dG:dC pair | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA polymerase IV, ... | | 著者 | Patra, A, Banerjee, S, Stone, M.P, Egli, M. | | 登録日 | 2014-11-18 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis for Error-Free Bypass of the 5-N-Methylformamidopyrimidine-dG Lesion by Human DNA Polymerase eta and Sulfolobus solfataricus P2 Polymerase IV.
J.Am.Chem.Soc., 137, 2015
|
|
4RW1
 
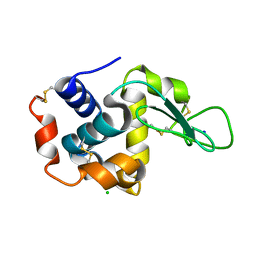 | | Hen egg-white lysozyme structure from a spent-beam experiment at LCLS: original beam | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Boutet, S, Foucar, L, Botha, S, Doak, R.B, Koglin, J.E, Messerschmidt, M, Nass, K, Schlichting, I, Shoeman, R, Williams, G.J. | | 登録日 | 2014-12-01 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization and use of the spent beam for serial operation of LCLS.
J.SYNCHROTRON RADIAT., 22, 2015
|
|
7WXX
 
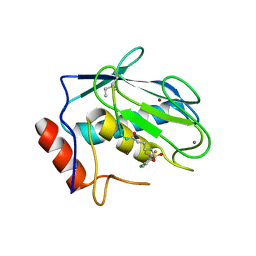 | | Crystal structure of human MMP-7 in complex with inhibitor | | 分子名称: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | 著者 | Kamitani, M, Oka, Y, Tabuse, H. | | 登録日 | 2022-02-15 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Highly Potent and Selective Matrix Metalloproteinase-7 Inhibitors by Hybridizing the S1' Subsite Binder with Short Peptides.
J.Med.Chem., 65, 2022
|
|
3BQ9
 
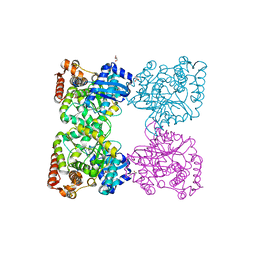 | | Crystal structure of predicted nucleotide-binding protein from Idiomarina baltica OS145 | | 分子名称: | GLYCEROL, Predicted Rossmann fold nucleotide-binding domain-containing protein, SULFATE ION | | 著者 | Patskovsky, Y, Toro, R, Meyer, A.J, Dickey, M, Eberle, M, Koss, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-12-19 | | 公開日 | 2008-01-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Predicted Nucleotide-Binding Protein from Idiomarina baltica.
To be Published
|
|
10MH
 
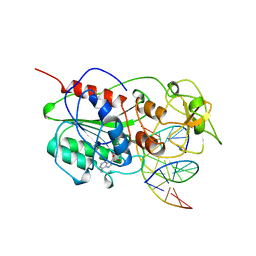 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH ADOHCY AND HEMIMETHYLATED DNA CONTAINING 5,6-DIHYDRO-5-AZACYTOSINE AT THE TARGET | | 分子名称: | DNA (5'-D(P*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*5NCP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), ... | | 著者 | Sheikhnejad, G, Brank, A, Christman, J.K, Goddard, A, Alvarez, E, Ford Junior, H, Marquez, V.E, Marasco, C.J, Sufrin, J.R, O'Gara, M, Cheng, X. | | 登録日 | 1998-08-10 | | 公開日 | 1999-02-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-azacytosine.
J.Mol.Biol., 285, 1999
|
|
116D
 
 | |
