4MGB
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with TCBPA | | 分子名称: | 4,4'-propane-2,2-diylbis(2,6-dichlorophenol), Estrogen receptor, GLYCEROL, ... | | 著者 | Delfosse, V, Grimaldi, M, Bourguet, W. | | 登録日 | 2013-08-28 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
3M5V
 
 | | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | 著者 | Kim, Y, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-03-13 | | 公開日 | 2010-04-28 | | 最終更新日 | 2012-02-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Dihydrodipicolinate Synthase from Campylobacter jejuni
To be Published
|
|
6F6D
 
 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | 分子名称: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | 著者 | Jones, S.E, Olsen, L, Gajhede, M. | | 登録日 | 2017-12-05 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.81810677 Å) | | 主引用文献 | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
7ZYR
 
 | | Compound 20 Bound to CK2alpha | | 分子名称: | 5-bromanyl-6-chloranyl-3-(1~{H}-pyrrol-2-ylmethyl)-1~{H}-indole, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Brear, P, Hyvonen, M. | | 登録日 | 2022-05-25 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A fragment-based approach leading to the discovery of inhibitors of CK2 alpha with a novel mechanism of action.
Rsc Med Chem, 13, 2022
|
|
1NH0
 
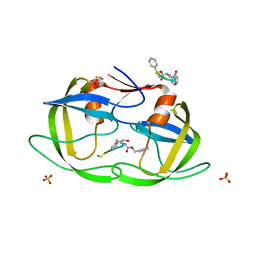 | | 1.03 A structure of HIV-1 protease: inhibitor binding inside and outside the active site | | 分子名称: | BETA-MERCAPTOETHANOL, PROTEASE RETROPEPSIN, SULFATE ION, ... | | 著者 | Brynda, J, Rezacova, P, Fabry, M, Horejsi, M, Hradilek, M, Soucek, M, Konvalinka, J, Sedlacek, J. | | 登録日 | 2002-12-18 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | A Phenylnorstatine Inhibitor Binding to HIV-1 Protease: Geometry,
Protonation, and Subsite-Pocket Interactions Analyzed at Atomic Resolution
J.Med.Chem., 47, 2004
|
|
1KER
 
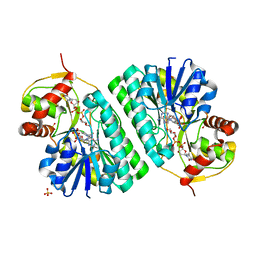 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | 分子名称: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | 登録日 | 2001-11-17 | | 公開日 | 2002-01-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6JGC
 
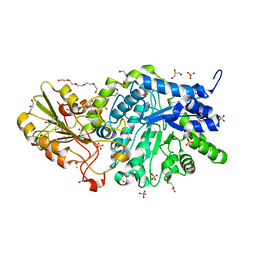 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with glucose. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-02-13 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
2MQF
 
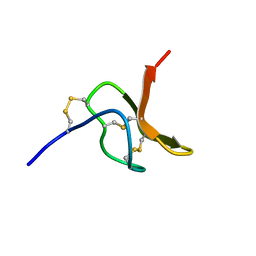 | |
1KFM
 
 | |
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | 分子名称: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | 著者 | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | 登録日 | 2013-08-29 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
6JGS
 
 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-02-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
1KFW
 
 | |
8A8U
 
 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
2BWJ
 
 | | Structure of adenylate kinase 5 | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENYLATE KINASE 5, CHLORIDE ION, ... | | 著者 | Bunkoczi, G, Filippakopoulos, P, Fedorov, O, Jansson, A, Longman, E, Ugochukwu, E, Knapp, S, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | 登録日 | 2005-07-14 | | 公開日 | 2005-07-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of Adenylate Kinase 5
To be Published
|
|
4YOP
 
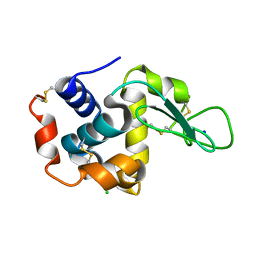 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE LYSOZYME | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Sugahara, M, Nakane, T, Suzuki, M, Nango, E. | | 登録日 | 2015-03-12 | | 公開日 | 2015-12-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Native sulfur/chlorine SAD phasing for serial femtosecond crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1KGL
 
 | | Solution structure of cellular retinol binding protein type-I in complex with all-trans-retinol | | 分子名称: | CELLULAR RETINOL-BINDING PROTEIN TYPE I, RETINOL | | 著者 | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | 登録日 | 2001-11-27 | | 公開日 | 2002-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and Backbone Dynamics of Apo- and Holo-cellular Retinol-binding
Protein in Solution.
J.Biol.Chem., 277, 2002
|
|
2MY8
 
 | |
5ABS
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL COILED-COIL DOMAIN OF CIN85 IN SPACE GROUP P321 | | 分子名称: | SH3 DOMAIN-CONTAINING KINASE-BINDING PROTEIN 1, ZINC ION | | 著者 | Wong, L, Habeck, M, Griesinger, C, Becker, S. | | 登録日 | 2015-08-07 | | 公開日 | 2016-07-13 | | 最終更新日 | 2019-02-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | The Adaptor Protein Cin85 Assembles Intracellular Signaling Clusters for B Cell Activation.
Sci.Signal., 9, 2016
|
|
8A8V
 
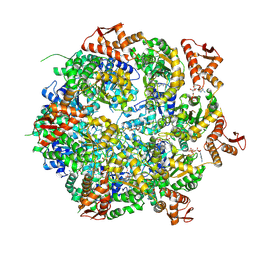 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8W
 
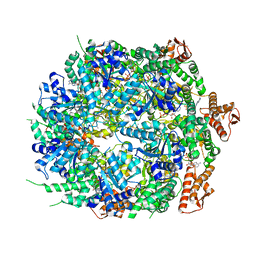 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | 著者 | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | 登録日 | 2022-06-24 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.29 Å) | | 主引用文献 | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
6F3C
 
 | | The cytotoxic [Pt(H2bapbpy)] platinum complex interacting with the CGTACG hexamer | | 分子名称: | DNA (5'-D(*GP*TP*AP*CP*G)-3'), MAGNESIUM ION, [Pt(H2bapbpy)] platinum | | 著者 | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Papi, F. | | 登録日 | 2017-11-28 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5'-d(CGTACG)-3' by a Mononuclear Platinum Complex.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6JJW
 
 | | Crystal Structure of KIBRA and PTPN14 complex | | 分子名称: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | 登録日 | 2019-02-27 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
6FD4
 
 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 14 hours post crystallization | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Adenine phosphoribosyltransferase, MAGNESIUM ION | | 著者 | Nioche, P, Huyet, J, Ozeir, M. | | 登録日 | 2017-12-21 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
4LUE
 
 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | 分子名称: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-25 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M9F
 
 | | Dengue virus NS2B-NS3 protease A125C variant at pH 8.5 | | 分子名称: | NS2B-NS3 protease | | 著者 | Yildiz, M, Ghosh, S, Bell, J.A, Sherman, W, Hardy, J.A. | | 登録日 | 2013-08-14 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus.
Acs Chem.Biol., 8, 2013
|
|
