6IAH
 
 | | Phosphatase Tt82 from Thermococcus thioreducens | | 分子名称: | CHLORIDE ION, Hydrolase, MAGNESIUM ION | | 著者 | Havlickova, P, Brinsa, V, Brynda, J, Pachl, P, Prudnikova, T, Mesters, J.R, Kascakova, B, Kuty, M, Pusey, M.L, Ng, J.D, Rezacova, P, Smatanova, I.K. | | 登録日 | 2018-11-26 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A novel structurally characterized haloacid dehalogenase superfamily phosphatase from Thermococcus thioreducens with diverse substrate specificity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | 分子名称: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rapp, M, Shapiro, L, Ho, D.D. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
6FX4
 
 | |
8OGJ
 
 | | Structure of Candida albicans 80S ribosome in complex with mefloquine | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | 著者 | Kolosova, O, Zgadzay, Y, Stetsenko, A, Guskov, A, Yusupov, M. | | 登録日 | 2023-03-20 | | 公開日 | 2024-10-02 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
5H6S
 
 | | Crystal structure of Hydrazidase S179A mutant complexed with a substrate | | 分子名称: | 4-oxidanylbenzohydrazide, Amidase | | 著者 | Akiyama, T, Ishii, M, Takuwa, A, Oinuma, K, Sasaki, Y, Takaya, N, Yajima, S. | | 登録日 | 2016-11-15 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate recognition of hydrazidase isolated from Microbacterium sp. strain HM58-2, which catalyzes acylhydrazide compounds as its sole carbon source
Biochem. Biophys. Res. Commun., 482, 2017
|
|
8OH6
 
 | | Crystal structure of the Candida albicans 80S ribosome in complex with Paromomycin (500umol) | | 分子名称: | 18S, 25S (gene name XR_002086444.1), 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | 著者 | Kolosova, O, Zgadzay, Y, Yusupov, M. | | 登録日 | 2023-03-20 | | 公開日 | 2024-10-02 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
5I5O
 
 | |
8PQX
 
 | | p97 (VCP) mutant - F539A state III | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | 著者 | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | 登録日 | 2023-07-12 | | 公開日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | p97 (VCP) mutant - F539A state III
To Be Published
|
|
5HA1
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with retinylamine | | 分子名称: | (2~{E},4~{E},6~{E},8~{E})-3,7-dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraen-1-amine, Retinol-binding protein 1 | | 著者 | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | 登録日 | 2015-12-29 | | 公開日 | 2016-03-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
6XGF
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-06-17 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8PDE
 
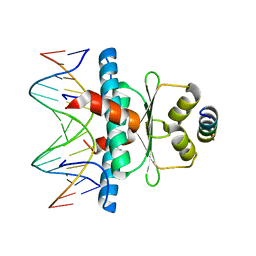 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC4 deacetylase binding motif | | 分子名称: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), HDAC4 (histone deacetylase 4) binding motif peptide:GSGEVKMKLQEFVLNKK, ... | | 著者 | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | 登録日 | 2023-06-12 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Spacer | | 著者 | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | 登録日 | 2016-02-22 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5HL7
 
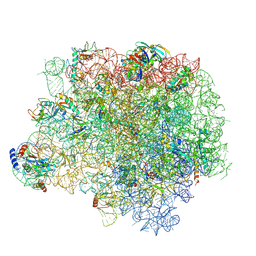 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lefamulin | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Eyal, Z, Matzov, D, Krupkin, M, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | 登録日 | 2016-01-14 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism.
Sci Rep, 6, 2016
|
|
8PPS
 
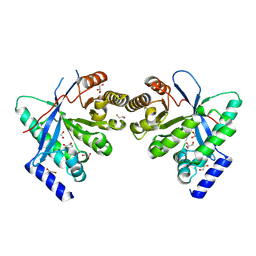 | | Dimeric RbdA EAL, in apo state | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, EAL domain-containing protein, ... | | 著者 | Cordery, C.R, Maly, M, Walsh, M.A, Tews, I. | | 登録日 | 2023-07-08 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Phosphodiesterase activation in the biofilm dispersal protein RbdA and relationship to the biofilm formation protein PA2072 of similar architecture
To Be Published
|
|
2Y6R
 
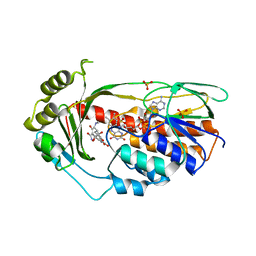 | | Structure of the TetX monooxygenase in complex with the substrate 7- chlortetracycline | | 分子名称: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | 登録日 | 2011-01-25 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
8PCH
 
 | | CRYSTAL STRUCTURE OF PORCINE CATHEPSIN H DETERMINED AT 2.1 ANGSTROM RESOLUTION: LOCATION OF THE MINI-CHAIN C-TERMINAL CARBOXYL GROUP DEFINES CATHEPSIN H AMINOPEPTIDASE FUNCTION | | 分子名称: | CATHEPSIN H, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Guncar, G, Podobnik, M, Pungercar, J, Strukelj, B, Turk, V, Turk, D. | | 登録日 | 1997-11-07 | | 公開日 | 1998-12-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of porcine cathepsin H determined at 2.1 A resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function.
Structure, 6, 1998
|
|
4JMA
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-Fluorocatechol | | 分子名称: | 3-FLUOROBENZENE-1,2-DIOL, Cytochrome c peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Boyce, S.E, Fischer, M, Fish, I. | | 登録日 | 2013-03-13 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
6XKT
 
 | | R. capsulatus cyt bc1 with both FeS proteins in c position (CIII2 c-c) | | 分子名称: | Cytochrome b, Cytochrome c1, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Steimle, S, Van Eeuwen, T, Ozturk, Y, Kim, H.J, Braitbard, M, Selamoglu, N, Garcia, B.A, Schneidman-Duhovny, D, Murakami, K, Daldal, F. | | 登録日 | 2020-06-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2021-03-03 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Nat Commun, 12, 2021
|
|
5HSV
 
 | | X-Ray structure of a CypA-Alisporivir complex at 1.5 angstrom resolution | | 分子名称: | Alisporivir, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Dujardin, M, Bouckaert, J, Rucktooa, P, Hanoulle, X. | | 登録日 | 2016-01-26 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray structure of alisporivir in complex with cyclophilin A at 1.5 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6XG3
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, at room temperature | | 分子名称: | CHLORIDE ION, Non-structural protein 3, PHOSPHATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5HTP
 
 | |
3D75
 
 | | Crystal structure of a pheromone binding protein mutant D35N, from Apis mellifera, at pH 5.5 | | 分子名称: | N-BUTYL-BENZENESULFONAMIDE, Pheromone-binding protein ASP1 | | 著者 | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | 登録日 | 2008-05-20 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
2Y6Q
 
 | | Structure of the TetX monooxygenase in complex with the substrate 7- Iodtetracycline | | 分子名称: | 7-IODOTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Volkers, G, Palm, G.J, Weiss, M.S, Wright, G.D, Hinrichs, W. | | 登録日 | 2011-01-25 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
5IHF
 
 | |
7SUC
 
 | |
