5R3N
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 47, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.059 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RC7
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library F11a | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCN
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 08 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RD3
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 25 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDK
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 44 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RE0
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 57 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1MO0
 
 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
2A57
 
 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
3P9O
 
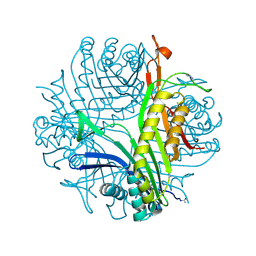 | | Aerobic ternary complex of urate oxidase with azide and chloride | | Descriptor: | AZIDE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gabison, L, Colloc'H, N, El Hajji, M, Castro, B, Chiadmi, M, Prange, T. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Azide and Cyanide Show Different Inhibition Modes to Urate Oxidase
To be Published
|
|
4NZW
 
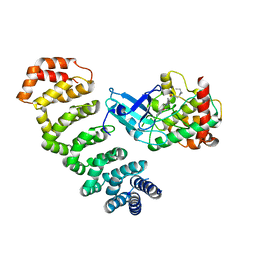 | | Crystal Structure of STK25-MO25 Complex | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | Authors: | Feng, M, Hao, Q, Zhou, Z.C. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
2A59
 
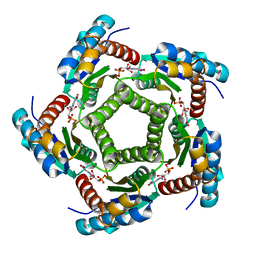 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2D55
 
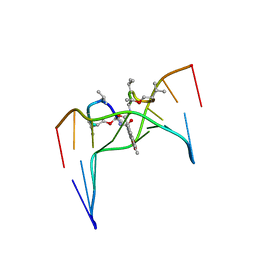 | | Structural, physical and biological characteristics of RNA.DNA binding agent N8-actinomycin D | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Shinomiya, M, Chu, W, Carlson, R.G, Weaver, R.F, Takusagawa, F. | | Deposit date: | 1995-05-01 | | Release date: | 1995-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 2:1 Complex between D(Gaagcttc) and the Anticancer Drug Actinomycin D.
J.Mol.Biol., 225, 1992
|
|
3PE1
 
 | | Crystal structure of human protein kinase CK2 alpha subunit in complex with the inhibitor CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Papinutto, E, Lolli, G, Pierre, F, Haddach, M, Ryckman, D.M. | | Deposit date: | 2010-10-25 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer.
Biochemistry, 50, 2011
|
|
1MGT
 
 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | Descriptor: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | Authors: | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
3PE2
 
 | | Crystal structure of human protein kinase CK2 in complex with the inhibitor CX-5011 | | Descriptor: | 5-[(3-ethynylphenyl)amino]pyrimido[4,5-c]quinoline-8-carboxylic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Battistutta, R, Papinutto, E, Lolli, G, Pierre, F, Haddach, M, Ryckman, D.M. | | Deposit date: | 2010-10-25 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer.
Biochemistry, 50, 2011
|
|
1TMX
 
 | | Crystal structure of hydroxyquinol 1,2-dioxygenase from Nocardioides Simplex 3E | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Ferraroni, M, Travkin, V.M, Seifert, J, Schlomann, M, Golovleva, L, Scozzafava, A, Briganti, F. | | Deposit date: | 2004-06-11 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation.
J.Biol.Chem., 280, 2005
|
|
1TVZ
 
 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
3PJ8
 
 | | Structure of CDK2 in complex with a Pyrazolo[4,3-d]pyrimidine Bioisostere of Roscovitine. | | Descriptor: | (2R)-2-{[7-(benzylamino)-3-(propan-2-yl)-1H-pyrazolo[4,3-d]pyrimidin-5-yl]amino}butan-1-ol, Cell division protein kinase 2 | | Authors: | McNae, I.W, Jorda, R, Havlicek, L, Strnad, M, Voller, J, Walkinshaw, M.D, Krystof, V. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pyrazolo[4,3-d]pyrimidine Bioisostere of Roscovitine: Evaluation of a Novel Selective Inhibitor of Cyclin-Dependent Kinases with Antiproliferative Activity.
J.Med.Chem., 54, 2011
|
|
4O1G
 
 | | MTB adenosine kinase in complex with gamma-Thio-ATP | | Descriptor: | Adenosine kinase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SODIUM ION | | Authors: | Dostal, J, Brynda, J, Hocek, M, Pichova, I. | | Deposit date: | 2013-12-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
1MBI
 
 | | X-RAY CRYSTAL STRUCTURE OF THE FERRIC SPERM WHALE MYOGLOBIN: IMIDAZOLE COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | IMIDAZOLE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lionetti, C, Guanziroli, M.G, Frigerio, F, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1990-06-25 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of the ferric sperm whale myoglobin: imidazole complex at 2.0 A resolution.
J.Mol.Biol., 217, 1991
|
|
1LVE
 
 | |
2AV9
 
 | | Crystal Structure of the PA5185 protein from Pseudomonas Aeruginosa Strain PAO1. | | Descriptor: | SULFATE ION, Thioesterase | | Authors: | Chruszcz, M, Wang, S, Cymborowski, M, Kudritska, M, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-29 | | Release date: | 2005-10-18 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
5SUH
 
 | |
3PZC
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with Coenzyme A | | Descriptor: | ACETATE ION, Amino acid--[acyl-carrier-protein] ligase 1, COENZYME A, ... | | Authors: | Weygand-Durasevic, I, Luic, M, Mocibob, M, Ivic, N, Subasic, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Recognition by Novel Family of Amino Acid:[Carrier Protein] Ligases
Croatica Chemica Acta, 84, 2011
|
|
2B0P
 
 | | truncated S. aureus LytM, P212121 crystal form | | Descriptor: | ACETATE ION, CACODYLATE ION, Glycyl-glycine endopeptidase lytM, ... | | Authors: | Firczuk, M, Mucha, A, Bochtler, M. | | Deposit date: | 2005-09-14 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of active LytM.
J.Mol.Biol., 354, 2005
|
|
