2P3U
 
 | |
5RBQ
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library B06b | | Descriptor: | (1S)-1-(4-nitrophenyl)ethan-1-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5VI7
 
 | |
5RC5
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library F06b | | Descriptor: | (2R)-N-tert-butylpiperidine-2-carboxamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCK
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 05 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RD0
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 22 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDH
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 39 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RDO
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 01 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6KRO
 
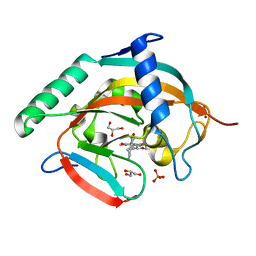 | | Tankyrase-2 in complex with RK-582 | | Descriptor: | 6-[(2S,6R)-2,6-dimethylmorpholin-4-yl]-4-fluoranyl-1-methyl-1'-(8-methyl-4-oxidanylidene-3,5,6,7-tetrahydropyrido[2,3-d]pyrimidin-2-yl)spiro[indole-3,4'-piperidine]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2019-08-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Discovery of an Orally Efficacious Spiroindolinone-Based Tankyrase Inhibitor for the Treatment of Colon Cancer.
J.Med.Chem., 63, 2020
|
|
1MG8
 
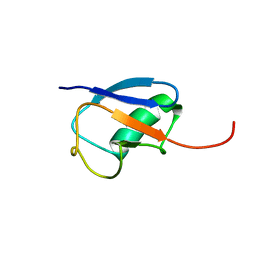 | | NMR structure of ubiquitin-like domain in murine Parkin | | Descriptor: | Parkin | | Authors: | Tashiro, M, Okubo, S, Shimotakahara, S, Hatanaka, H, Yasuda, H, Kainosho, M, Yokoyama, S, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ubiquitin-like domain in PARKIN: Gene product of familial Parkinson's disease.
J.Biomol.NMR, 25, 2003
|
|
1UU9
 
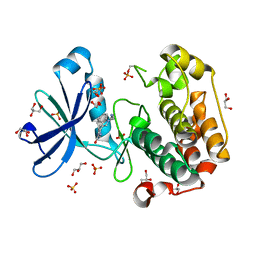 | | Structure of human PDK1 kinase domain in complex with BIM-3 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-[1-(3-AMINOPROPYL)-1H-INDOL-3-YL]-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-16 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
1UU7
 
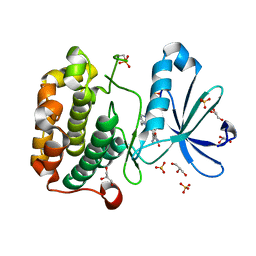 | | Structure of human PDK1 kinase domain in complex with BIM-2 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-(1-{2-[(2S)-1-METHYLPYRROLIDINYL]ETHYL}-1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-16 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
3OOH
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
5R12
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 16, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1UVR
 
 | | Structure of human PDK1 kinase domain in complex with BIM-8 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-[1-(3-AMINOPROPYL)-1H-INDOL-3-YL]-4-(1-METHYL-1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Garrido-Franco, M, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-01-22 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
5R1L
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 36, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1Y
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry D10, DMSO-free | | Descriptor: | 2-methyl-N-(4-methylphenyl)-L-alanine, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.038 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2B
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H03, DMSO-free | | Descriptor: | Endothiapepsin, N-[(benzyloxy)carbonyl]-N-methyl-L-alanine | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.019 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2P
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 13, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.179 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R34
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 28, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3J
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 43, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.077 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R40
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 60, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
1UU3
 
 | | Structure of human PDK1 kinase domain in complex with LY333531 | | Descriptor: | (9R)-9-[(DIMETHYLAMINO)METHYL]-6,7,10,11-TETRAHYDRO-9H,18H-5,21:12,17-DIMETHENODIBENZO[E,K]PYRROLO[3,4-H][1,4,13]OXADIA ZACYCLOHEXADECINE-18,20-DIONE, 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
5R0T
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 07, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R19
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 24, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
