1M1N
 
 | | Nitrogenase MoFe protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(7)-MO-S(9)-N CLUSTER, ... | | Authors: | Einsle, O, Tezcan, F.A, Andrade, S.L.A, Schmid, B, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-19 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor.
Science, 297, 2002
|
|
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
3UGT
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase - orthorhombic crystal form | | Descriptor: | Threonyl-tRNA synthetase, mitochondrial, ZINC ION | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1M7S
 
 | | Crystal Structure Analysis of Catalase CatF of Pseudomonas syringae | | Descriptor: | Catalase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Carpena, X, Soriano, M, Klotz, M.G, Duckworth, H.W, Donald, L.J, Melik-Adamyan, W, Fita, I, Loewen, P.C. | | Deposit date: | 2002-07-22 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Clade 1 catalase, CatF of Pseudomonas syringae, at 1.8 A resolution
Proteins, 50, 2003
|
|
3URK
 
 | | IspH in complex with propynyl diphosphate (1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, prop-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | Descriptor: | IODIDE ION, Putative uncharacterized protein | | Authors: | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UUA
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with bisphenol-AF | | Descriptor: | 4,4'-(1,1,1,3,3,3-hexafluoropropane-2,2-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UUN
 
 | |
1MN7
 
 | | NDP kinase mutant (H122G;N119S;F64W) in complex with aBAZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXY-THYMIDINE-5'-ALPHA BORANO TRIPHOSPHATE, MAGNESIUM ION, NDP kinase | | Authors: | gallois-montbrun, s, schneider, b, chen, y, giacomoni-fernandes, v, mulard, l, morera, s, janin, j, deville-bonne, d, veron, m. | | Deposit date: | 2002-09-05 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation
J.BIOL.CHEM., 277, 2002
|
|
1MG6
 
 | |
3UWQ
 
 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
1MOX
 
 | | Crystal Structure of Human Epidermal Growth Factor Receptor (residues 1-501) in complex with TGF-alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garrett, T.P.J, McKern, N.M, Lou, M, Elleman, T.C, Adams, T.E, Lovrecz, G.O, Zhu, H.-J, Walker, F, Frenkel, M.J, Hoyne, P.A, Jorissen, R.N, Nice, E.C, Burgess, A.W, Ward, C.W. | | Deposit date: | 2002-09-10 | | Release date: | 2003-09-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Truncated Epidermal Growth Factor Receptor Extracellular Domain Bound to Transforming Growth Factor alpha
Cell(Cambridge,Mass.), 110, 2002
|
|
1MH6
 
 | | Solution Structure of the Transposon Tn5-encoding Bleomycin-binding Protein, BLMT | | Descriptor: | BLEOMYCIN RESISTANCE PROTEIN | | Authors: | Kumagai, T, Ohtani, K, Tsuboi, Y, Koike, T, Sugiyama, M. | | Deposit date: | 2002-08-19 | | Release date: | 2003-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transposon Tn5-encoding bleomycin-binding protein complexed with an activated bleomycin analogue.
To be published
|
|
3UME
 
 | | Structure of pB intermediate of Photoactive yellow protein (PYP) at pH 7 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Tripathi, S, Srajer, V, Purwar, N, Henning, R, Schmidt, M. | | Deposit date: | 2011-11-13 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | pH Dependence of the Photoactive Yellow Protein Photocycle Investigated by Time-Resolved Crystallography.
Biophys.J., 102, 2012
|
|
3UMO
 
 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with Potassium | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Garratt, R.C, Babul, J. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | A Ribokinase Family Conserved Monovalent Cation Binding Site Enhances the MgATP-induced Inhibition in E. coli Phosphofructokinase-2
Biophys.J., 105, 2013
|
|
1LX8
 
 | | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein | | Descriptor: | Excisionase | | Authors: | Sam, M.D, Papagiannis, C, Connolly, K.M, Corselli, L, Iwahara, J, Lee, J, Phillips, M, Wojciak, J.M, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2002-06-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein
J.Mol.Biol., 324, 2002
|
|
1LZD
 
 | | DISSECTION OF PROTEIN-CARBOHYDRATE INTERACTIONS IN MUTANT HEN EGG-WHITE LYSOZYME COMPLEXES AND THEIR HYDROLYTIC ACTIVITY | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Maenaka, K, Matsushima, M, Song, H, Watanabe, K, Kumagai, I. | | Deposit date: | 1995-02-10 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissection of protein-carbohydrate interactions in mutant hen egg-white lysozyme complexes and their hydrolytic activity.
J.Mol.Biol., 247, 1995
|
|
1LZB
 
 | | DISSECTION OF PROTEIN-CARBOHYDRATE INTERACTIONS IN MUTANT HEN EGG-WHITE LYSOZYME COMPLEXES AND THEIR HYDROLYTIC ACTIVITY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEN EGG WHITE LYSOZYME | | Authors: | Maenaka, K, Matsushima, M, Song, H, Watanabe, K, Kumagai, I. | | Deposit date: | 1995-02-10 | | Release date: | 1995-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dissection of protein-carbohydrate interactions in mutant hen egg-white lysozyme complexes and their hydrolytic activity.
J.Mol.Biol., 247, 1995
|
|
1LZS
 
 | |
1LZQ
 
 | | Crystal structure of the complex of mutant HIV-1 protease (A71V, V82T, I84V) with an ethylenamine peptidomimetic inhibitor BOC-PHE-PSI[CH2CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | BETA-MERCAPTOETHANOL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Hasek, J, Dohnalek, J, Petrokova, H, Buchtelova, E, Soucek, M, Majer, P, Uhlikova, T, Konvalinka, J. | | Deposit date: | 2002-06-11 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Ethylenamine Inhibitor Binds Tightly to Both Wild Type and Mutant HIV-1 Proteases. Structure and Energy Study
J.Med.Chem., 46, 2003
|
|
1M11
 
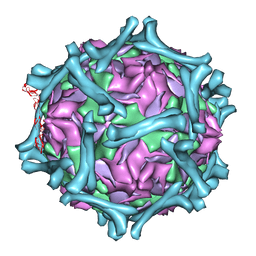 | | structural model of human decay-accelerating factor bound to echovirus 7 from cryo-electron microscopy | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | He, Y, Lin, F, Chipman, P.R, Bator, C.M, Baker, T.S, Shoham, M, Kuhn, R.J, Medof, M.E, Rossmann, M.G. | | Deposit date: | 2002-06-17 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3V1X
 
 | |
3V2J
 
 | |
1M2W
 
 | | Pseudomonas fluorescens mannitol 2-dehydrogenase ternary complex with NAD and D-mannitol | | Descriptor: | D-MANNITOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-06-25 | | Release date: | 2002-11-15 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pseudomonas fluorescens Mannitol 2-Dehydrogenase Binary and Ternary Complexes. Specificity and Catalytic Mechanism
J.Biol.Chem., 277, 2002
|
|
3V4A
 
 | | Structure of ar lbd with activator peptide and sarm inhibitor 2 | | Descriptor: | (5R)-3-(3,4-dichlorophenyl)-5-(4-hydroxyphenyl)-1,5-dimethyl-2-thioxoimidazolidin-4-one, Androgen receptor, SULFATE ION | | Authors: | Nique, F, Hebbe, S, Peixoto, C, Annoot, D, Lefrancois, J.-M, Duval, E, Michoux, L, Triballeau, N, Lemoullec, J.M, Mollat, P, Thauvin, M, Prange, T, Minet, D, Clement-Lacroix, P, Robin-Jagerschmidt, C, Fleury, D, Guedin, D, Deprez, P. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of diarylhydantoins as new selective androgen receptor modulators.
J.Med.Chem., 55, 2012
|
|
