3BHO
 
 | |
3BID
 
 | | Crystal structure of the NMB1088 protein from Neisseria meningitidis. Northeast Structural Genomics Consortium target MR91 | | Descriptor: | UPF0339 protein NMB1088 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, L, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-18 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the NMB1088 protein from Neisseria meningitidis.
To be Published
|
|
7WDB
 
 | | Human TRPC5 channel in complex with riluzole | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, CALCIUM ION, ... | | Authors: | Chen, L, Wei, M, Yang, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-04-13 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural identification of riluzole-binding site on human TRPC5.
Cell Discov, 8, 2022
|
|
3B96
 
 | | Structural Basis for Substrate Fatty-Acyl Chain Specificity: Crystal Structure of Human Very-Long-Chain Acyl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TETRADECANOYL-COA, Very long-chain specific acyl-CoA dehydrogenase | | Authors: | McAndrew, R.P, Wang, Y, Mohsen, A.W, He, M, Vockley, J, Kim, J.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for substrate fatty acyl chain specificity: crystal structure of human very-long-chain acyl-CoA dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3B9O
 
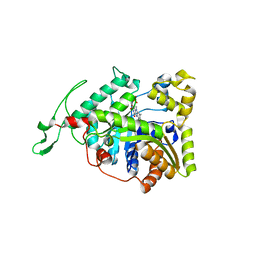 | | long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN | | Descriptor: | Alkane monooxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Yang, W, Xu, F, Bartlam, M, Rao, Z. | | Deposit date: | 2007-11-06 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase
J.Mol.Biol., 376, 2008
|
|
3BAU
 
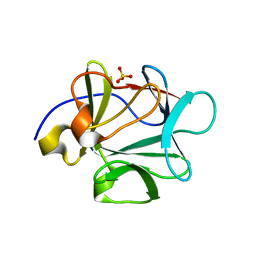 | |
7DY4
 
 | |
3BJD
 
 | | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Putative 3-oxoacyl-(acyl-carrier-protein) synthase | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-03 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-oxoacyl-(acyl-carrier-protein) synthase from Pseudomonas aeruginosa.
To be Published
|
|
3BB6
 
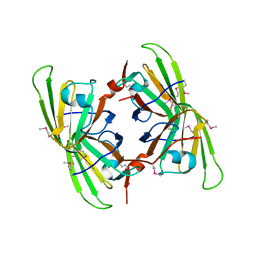 | | Crystal structure of the P64488 protein from E.coli (strain K12). Northeast Structural Genomics Consortium target ER596 | | Descriptor: | Uncharacterized protein yeaR, ZINC ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Janjua, H, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-09 | | Release date: | 2007-11-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray structure of the P64488 from E.coli (strain K12).
To be Published
|
|
7WOX
 
 | | PPARgamma antagonist (MMT-160)- PPARgamma LBD complex | | Descriptor: | N-[[5-(3-phenylprop-2-ynoylamino)-2-propoxy-phenyl]methyl]-4-pyrimidin-2-yl-benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshizawa, M, Aoyama, T, Itoh, T, Miyachi, H. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Arylalkynyl amide-type peroxisome proliferator-activated receptor gamma (PPAR gamma )-selective antagonists covalently bind to the PPAR gamma ligand binding domain with a unique binding mode.
Bioorg.Med.Chem.Lett., 64, 2022
|
|
7WJP
 
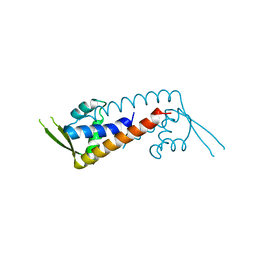 | | Structure of PadR-like protein from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Hong, M, Kim, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based molecular characterization of the LltR transcription factor from Listeria monocytogenes.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
3BDR
 
 | | Crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus. Northeast Structural Genomics Consortium target TeR13. | | Descriptor: | PHOSPHATE ION, Ycf58 protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of fatty acid-binding protein-like Ycf58 from Thermosynecoccus elongatus.
To be Published
|
|
3B2K
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3KIE
 
 | |
7EBT
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in glutathione-bound form | | Descriptor: | CALCIUM ION, GLUTATHIONE, Glutathione transferase | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
3BEW
 
 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
3BFB
 
 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with the 9-keto-2(E)-decenoic acid | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7VY3
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES LACKING PROTEIN-U | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Tani, K, Kanno, R, Kawamura, S, Kikuchi, R, Nagashima, K.V.P, Hall, M, Takahashi, A, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Asymmetric structure of the native Rhodobacter sphaeroides dimeric LH1-RC complex.
Nat Commun, 13, 2022
|
|
7EBW
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in desmethylglycitein and glutathione-bound form | | Descriptor: | 6,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
6VJ0
 
 | | Crystal structure of a chitin-binding protein from Moringa oleifera seeds (Mo-CBP4) | | Descriptor: | ACETATE ION, CHLORIDE ION, Chitin-binding protein Mo-CBP4 | | Authors: | Bezerra, E.H.S, Lopes, T.D.P, da Silva, F.M.S, Costa, H.P.S, Freire, V.N, Rocha, B.A.M, Sousa, D.O.B. | | Deposit date: | 2020-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a lectin from Moringa oleifera seeds with imflammatory activities
To Be Published
|
|
7EBU
 
 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in Daidzein- and glutathione-bound form | | Descriptor: | 7-hydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7VY2
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES DIMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Tani, K, Kanno, R, Kawamura, S, Kikuchi, R, Nagashima, K.V.P, Hall, M, Takahashi, A, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Asymmetric structure of the native Rhodobacter sphaeroides dimeric LH1-RC complex.
Nat Commun, 13, 2022
|
|
2B35
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
7E4D
 
 | | Crystal structure of PlDBR | | Descriptor: | Double Bond Reductase | | Authors: | Sugimoto, K, Senda, M, Senda, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploration and structure-based engineering of alkenal double bond reductases catalyzing the C alpha C beta double bond reduction of coniferaldehyde.
N Biotechnol, 68, 2022
|
|
7E1G
 
 | | Structure of MreB3 from Spiroplasma eriocheiris | | Descriptor: | Cell shape-determining protein MreB, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Takahashi, D, Miyata, M, Imada, K. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ATP-dependent polymerization dynamics of bacterial actin proteins involved in Spiroplasma swimming.
Open Biology, 12, 2022
|
|
