2CSZ
 
 | | Solution Structure of the RING domain of the Synaptotagmin-like protein 4 | | Descriptor: | Synaptotagmin-like protein 4, ZINC ION | | Authors: | Miyamoto, K, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RING domain of the Synaptotagmin-like protein 4
To be Published
|
|
3CDC
 
 | |
7QAO
 
 | | Three-dimensional structure of the PGAM5 C12S mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
3C9N
 
 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
2CKB
 
 | | STRUCTURE OF THE 2C/KB/DEV8 COMPLEX | | Descriptor: | ALPHA, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | Authors: | Garcia, K.C, Degano, M, Wilson, I.A. | | Deposit date: | 1998-01-14 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen.
Science, 279, 1998
|
|
3CDU
 
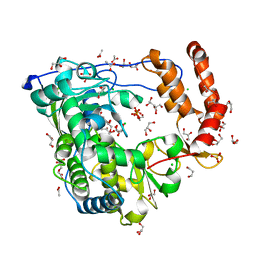 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
7QAM
 
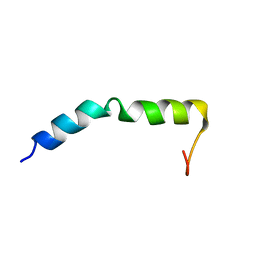 | | Three-dimensional structure of the PGAM5 WT TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
3CDY
 
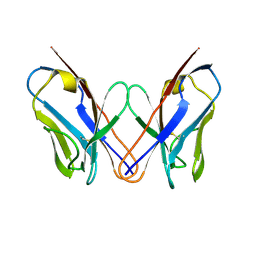 | | AL-09 H87Y, immunoglobulin light chain variable domain | | Descriptor: | IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Baden, E.M, Randles, E.G, Aboagye, A.K, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the role of mutations in amyloidogenesis.
J.Biol.Chem., 283, 2008
|
|
3CD0
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{2-[(4-fluorobenzyl)carbamoyl]-4-(4-fluorophenyl)-1-(1-methylethyl)-1H-imidazol-5-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
2CPJ
 
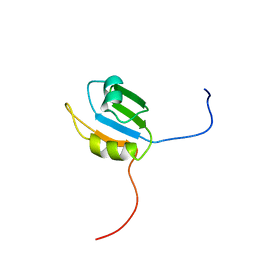 | | Solution structure of the N-terminal RNA recognition motif of NonO | | Descriptor: | Non-POU domain-containing octamer-binding protein | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA recognition motif of NonO
To be Published
|
|
3CD7
 
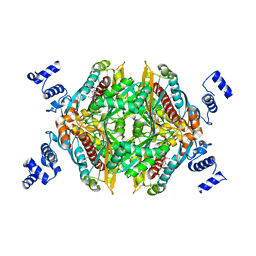 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
3CDB
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{3-[(4-carbamoylphenyl)sulfamoyl]-4,5-bis(4-fluorophenyl)-2-(1-methylethyl)-1H-pyrrol-1-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
2CQD
 
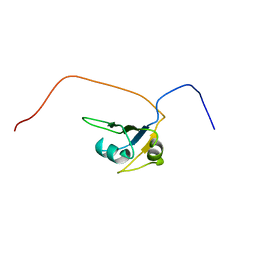 | | Solution Structure of the RNA recognition motif in RNA-binding region containing protein 1 | | Descriptor: | RNA-binding region containing protein 1 | | Authors: | Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RNA recognition motif in RNA-binding region containing protein 1
To be Published
|
|
7QH7
 
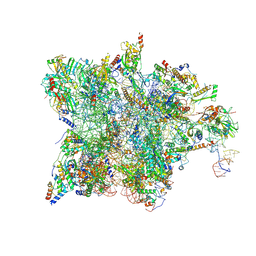 | | Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 4 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Rebelo-Guiomar, P, Pellegrino, S, Dent, K.C, Warren, A.J, Minczuk, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Nat Commun, 13, 2022
|
|
3CDL
 
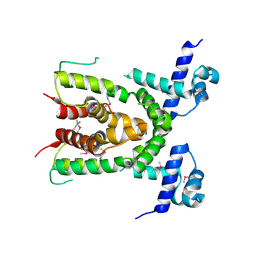 | |
7QRE
 
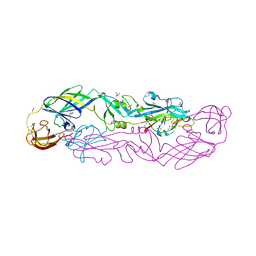 | | Structure of the hetero-tetramer complex between precursor membrane protein fragment (pr) and envelope protein (E) from tick-borne encephalitis virus | | Descriptor: | ACETATE ION, Envelope protein E, Genome polyprotein, ... | | Authors: | Vaney, M.C, Dellarole, M, Rey, F.A. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
3CDN
 
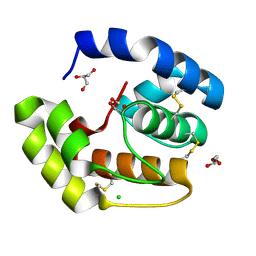 | | Crystal structure of a pheromone binding protein from Apis mellifera soaked at pH 4.0 | | Descriptor: | CHLORIDE ION, GLYCEROL, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7P0N
 
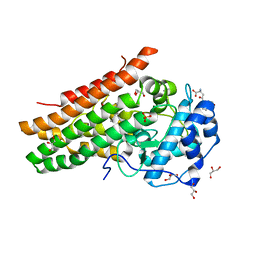 | | Crystal structure of L-Trp/Indoleamine 2,3-dioxygenagse 1 (hIDO1) complex with the JK-loop refined in the open conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Mirgaux, M, Wouters, J. | | Deposit date: | 2021-06-30 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Temporary Intermediates of L-Trp Along the Reaction Pathway of Human Indoleamine 2,3-Dioxygenase 1 and Identification of an Exo Site.
Int J Tryptophan Res, 14, 2021
|
|
3CAR
 
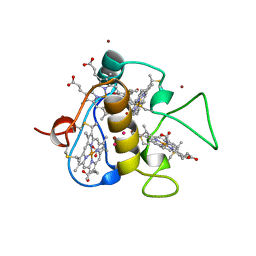 | | REDUCED STRUCTURE OF THE ACIDIC CYTOCHROME C3 FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | ARSENIC, CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Norager, S, Legrand, P, Pieulle, L, Hatchikian, C, Roth, M. | | Deposit date: | 1998-11-17 | | Release date: | 2000-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the oxidised and reduced acidic cytochrome c3from Desulfovibrio africanus.
J.Mol.Biol., 290, 1999
|
|
3CDZ
 
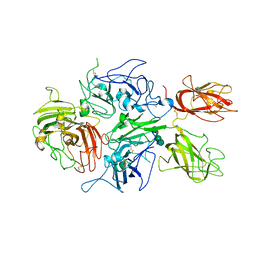 | | Crystal structure of human factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ngo, J.C, Huang, M, Roth, D.A, Furie, B.C, Furie, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex.
Structure, 16, 2008
|
|
3CDW
 
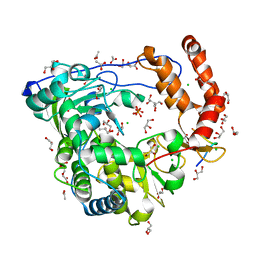 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
2CTO
 
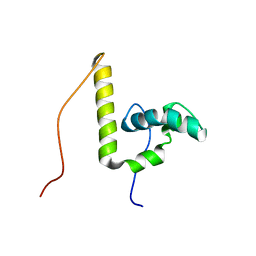 | | Solution structure of the HMG box like domain from human hypothetical protein FLJ14904 | | Descriptor: | novel protein | | Authors: | Tomizawa, T, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Kamatari, Y.O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG box like domain from human hypothetical protein FLJ14904
To be Published
|
|
2CNH
 
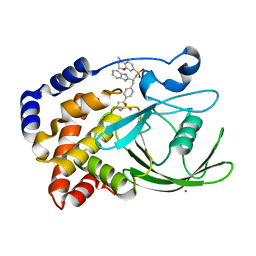 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | CALCIUM ION, N-[(1S)-1-(1H-BENZIMIDAZOL-2-YL)-2-{4-[(5S)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]PHENYL}ETHYL]-4-METHYL-3,4-DIHYDRO-2H-1,4-BENZOXAZINE-7-SULFONAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
3CDA
 
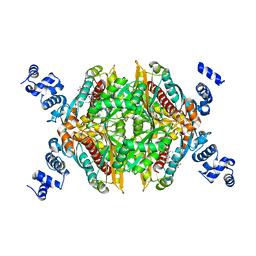 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-{3-(4-fluorophenyl)-1-(1-methylethyl)-4-phenyl-5-[(4-sulfamoylphenyl)carbamoyl]-1H-pyrrol-2-yl}-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
6HE6
 
 | |
