6ZBL
 
 | |
6U6R
 
 | |
8J4C
 
 | | YeeE(TsuA)-YeeD(TsuB) complex for thiosulfate uptake | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Spirochaeta thermophila YeeE(TsuA)-YeeD(TsuB),UPF0033 domain-containing protein, SirA-like domain-containing protein (chimera), ... | | Authors: | Ikei, M, Miyazaki, R, Monden, K, Naito, Y, Takeuchi, A, Takahashi, Y.S, Tanaka, Y, Ichikawa, M, Tsukazaki, T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | YeeD is an essential partner for YeeE-mediated thiosulfate uptake in bacteria and regulates thiosulfate ion decomposition.
Plos Biol., 22, 2024
|
|
6ZBG
 
 | |
4MO1
 
 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | Descriptor: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
6U6P
 
 | |
3ZBT
 
 | | Ferredoxin-NADP Reductase Mutant with SER 59 Replaced by ALA (S59A) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
3ZMU
 
 | | LSD1-CoREST in complex with PKSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
3KJL
 
 | | Sgf11:Sus1 complex | | Descriptor: | Protein SUS1, SAGA-associated factor 11 | | Authors: | Stewart, M, Ellisdon, A.M. | | Deposit date: | 2009-11-03 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1.
J.Biol.Chem., 285, 2010
|
|
8YJ7
 
 | | Characerization of a novel format scFvXVHH single-chain Biparatopic antibody against a metal binding protein, MtsA | | Descriptor: | Iron ABC transporter substrate-binding lipoprotein MtsA, ZINC ION, scFv13 | | Authors: | Ito, S, Nagatoishi, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2024-03-01 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of a novel format scFv×VHH single-chain biparatopic antibody against metal binding protein MtsA.
Protein Sci., 33, 2024
|
|
3ZKQ
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MU6
 
 | |
7QX3
 
 | | Structure of the transaminase TR2E2 with EOS | | Descriptor: | 2-azanylethyl hydrogen sulfate, Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7QX0
 
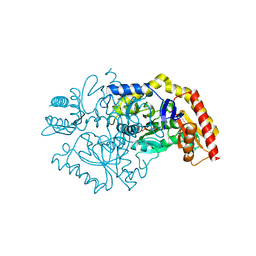 | | Transaminase Structure of Plurienzyme (Tr2E2) in complex with PLP | | Descriptor: | Aminotransferase TR2, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-26 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
2AF0
 
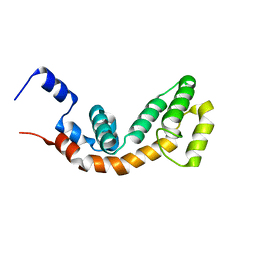 | | Structure of the Regulator of G-Protein Signaling Domain of RGS2 | | Descriptor: | Regulator of G-protein signaling 2 | | Authors: | Papagrigoriou, E, Johannson, C, Phillips, C, Smee, C, Elkins, J.M, Weigelt, J, Arrowsmith, C, Edwards, A, Sundstrom, M, Von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ZH0
 
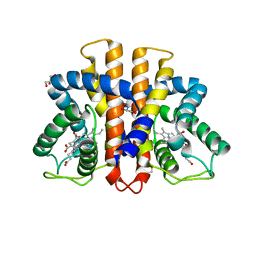 | | Functional and structural role of the N-terminal extension in Methanosarcina acetivorans protoglobin | | Descriptor: | FORMIC ACID, GLYCEROL, PROTOGLOBIN, ... | | Authors: | Ciaccio, C, Pesce, A, Tundo, G.R, Tilleman, L, Dewilde, S, Moens, L, Ascenzi, P, Bolognesi, M, Nardini, M, Coletta, M. | | Deposit date: | 2012-12-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Role of the N-Terminal Extension in Methanosarcina Acetivorans Protoglobin.
Biochim.Biophys.Acta, 1834, 2013
|
|
8ZNG
 
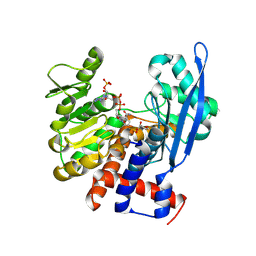 | |
285D
 
 | |
3ZN0
 
 | | LSD1-CoREST in complex with PRSFAA peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
6LBH
 
 | |
3C8G
 
 | | Crystal structure of a possible transciptional regulator YggD from Shigella flexneri 2a str. 2457T | | Descriptor: | ACETATE ION, Putative transcriptional regulator | | Authors: | Tan, K, Borovilos, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
8ZNE
 
 | |
8ZNB
 
 | |
6U6S
 
 | |
