6J26
 
 | | Crystal structure of the branched-chain polyamine synthase from Thermococcus kodakarensis (Tk-BpsA) in complex with N4-bis(aminopropyl)spermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
8WTC
 
 | |
7UNO
 
 | | Thiol-disulfide oxidoreductase TsdA, C129S mutant, from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Reardon-Robinson, M, Nguyen, M.T, Sanchez, B, Ton-That, H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-11 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thiol-disulfide oxidoreductase in Corynebacterium diphtheriae
To Be Published
|
|
7PQP
 
 | | tau-microtubule structural ensemble based on CryoEM data | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Isoform Tau-F of Microtubule-associated protein tau, ... | | Authors: | Brotzakis, Z.F, Vendruscolo, M. | | Deposit date: | 2021-09-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A Structural Ensemble of a Tau-Microtubule Complex Reveals Regulatory Tau Phosphorylation and Acetylation Mechanisms.
Acs Cent.Sci., 7, 2021
|
|
8WTB
 
 | |
1ENN
 
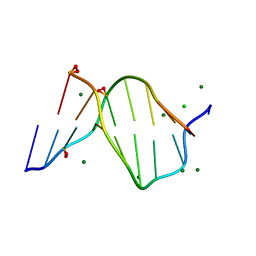 | | SOLVENT ORGANIZATION IN AN OLIGONUCLEOTIDE CRYSTAL: THE STRUCTURE OF D(GCGAATTCG)2 AT ATOMIC RESOLUTION | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*GP*AP*AP*TP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Soler-Lopez, M, Malinina, L, Subirana, J.A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution.
J.Biol.Chem., 275, 2000
|
|
8WY0
 
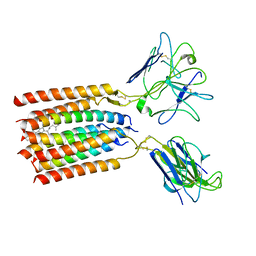 | | T cell receptor delta 2 gamma 9 with F283A, F290A, and F291A | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
6J65
 
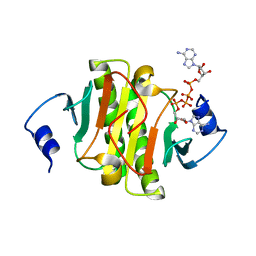 | | Crystal structure of human HINT1 mutant complexing with AP4A II | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
8WXE
 
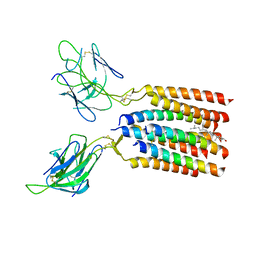 | | Vgamma5Vdelta1 EH TCR-CD3 complex | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 1,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 5,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
7PQC
 
 | | tau-microtubule structural ensemble based on CryoEM data | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Isoform Tau-F of Microtubule-associated protein tau, ... | | Authors: | Brotzakis, Z.F, Vendruscolo, M. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A Structural Ensemble of a Tau-Microtubule Complex Reveals Regulatory Tau Phosphorylation and Acetylation Mechanisms.
Acs Cent.Sci., 7, 2021
|
|
8WYI
 
 | | T cell receptor delta 2 gamma 9 with TCRD TM domain chimera of TRAC | | Descriptor: | Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1,T cell receptor gamma variable 9,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
1E5A
 
 | | Structure of human transthyretin complexed with bromophenols: a new mode of binding | | Descriptor: | 2,4,6-TRIBROMOPHENOL, TRANSTHYRETIN | | Authors: | Ghosh, M, Meerts, I.A.T.M, Cook, A, Bergman, A, Brouwer, A, Johnson, L.N. | | Deposit date: | 2000-07-20 | | Release date: | 2000-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Human Transthyretin Complexed with Bromophenols : A New Mode of Binding
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4DPV
 
 | | PARVOVIRUS/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Chapman, M.S, Rossmann, M.G. | | Deposit date: | 1996-02-01 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
1EQK
 
 | | SOLUTION STRUCTURE OF ORYZACYSTATIN-I, A CYSTEINE PROTEINASE INHIBITOR OF THE RICE, ORYZA SATIVA L. JAPONICA | | Descriptor: | ORYZACYSTATIN-I | | Authors: | Nagata, K, Kudo, N, Abe, K, Arai, S, Tanokura, M. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica.
Biochemistry, 39, 2000
|
|
8WMW
 
 | | The structure of PSI-11CAC at the stationary growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
8WMV
 
 | | The structure of PSI-14CAC complex at stationary growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
8WMJ
 
 | | structure of PSI-11CAC complex at Logrithmic growth phase | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
1EYR
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | Descriptor: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE, CYTIDINE-5'-DIPHOSPHATE | | Authors: | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | Deposit date: | 2000-05-08 | | Release date: | 2001-02-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
8WM6
 
 | | The structure of PSI-CAC(L-14)of R.salina at 2.7 angstroms resolution | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhang, S.M, Si, L, Li, M. | | Deposit date: | 2023-10-03 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Growth phase-dependent reorganization of cryptophyte photosystem I antennae.
Commun Biol, 7, 2024
|
|
1EEP
 
 | | 2.4 A RESOLUTION CRYSTAL STRUCTURE OF BORRELIA BURGDORFERI INOSINE 5'-MONPHOSPHATE DEHYDROGENASE IN COMPLEX WITH A SULFATE ION | | Descriptor: | INOSINE 5'-MONOPHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | McMillan, F.M, Cahoon, M, White, A, Hedstrom, L, Petsko, G.A, Ringe, D. | | Deposit date: | 2000-02-01 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of Borrelia burgdorferi inosine 5'-monophosphate dehydrogenase: evidence of a substrate-induced hinged-lid motion by loop 6.
Biochemistry, 39, 2000
|
|
7PRG
 
 | | Joint X-ray/neutron room temperature structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | Descriptor: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | Authors: | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | Deposit date: | 2021-09-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
8WUT
 
 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | Authors: | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2023-10-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
3CM2
 
 | | Crystal Structure of XIAP BIR3 domain in complex with a Smac-mimetic compound, Smac010 | | Descriptor: | (3S,6S,7R,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-7-(aminomethyl)-N-(diphenylmethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Cossu, F, Mastrangelo, E, Milani, M. | | Deposit date: | 2008-03-20 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane smac mimetics. Structure, activity, and recognition principles.
J.Mol.Biol., 384, 2008
|
|
6JMT
 
 | | Crystal structure of GIT/PIX complex | | Descriptor: | ARF GTPase-activating protein GIT2, ZINC ION, beta PIX | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
