6GPY
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with AMPPNP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
3QD7
 
 | | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli | | Descriptor: | Uncharacterized protein ydaL | | Authors: | Gui, W.J, Qu, Q.H, Chen, Y.Y, Wang, M, Zhang, X.E, Bi, L.J, Jiang, T. | | Deposit date: | 2011-01-18 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YdaL, a stand-alone small MutS-related protein from Escherichia coli.
J.Struct.Biol., 174, 2011
|
|
6GQ5
 
 | |
4IRS
 
 | | Structure of the mouse CD1d-PyrC-alpha-GalCer-iNKT TCR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nemcovic, M, Zajonc, D.M. | | Deposit date: | 2013-01-15 | | Release date: | 2013-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enhanced TCR footprint by a novel glycolipid increases NKT-dependent tumor protection.
J.Immunol., 191, 2013
|
|
6GR3
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with AMPPNP | | Descriptor: | AMP PHOSPHORAMIDATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
6H6E
 
 | | PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) | | Descriptor: | TcdA1, TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6GRN
 
 | | CELLOBIOHYDROLASE I (CEL7A) FROM Trichoderma reesei with S-dihydroxypropranolol in the active site | | Descriptor: | 2-[[(2~{S})-3-naphthalen-1-yloxy-2-oxidanyl-propyl]amino]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Sandgren, M, Fagerstrom, A, Widmalm, G, Stahlberg, J. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Enantioselective Binding of Propranolol and Analogues Thereof to Cellobiohydrolase Cel7A.
Chemistry, 24, 2018
|
|
3QF6
 
 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | Deposit date: | 2011-01-21 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.85 Å) | | Cite: | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
6GSF
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2018-06-14 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
4O7G
 
 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 40 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6H76
 
 | | SiaP in complex with Neu5Ac (RT) | | Descriptor: | CESIUM ION, CHLORIDE ION, N-acetyl-beta-neuraminic acid, ... | | Authors: | Fischer, M, Darby, J.F, Brannigan, J.A, Turkenburg, J, Hubbard, R.E. | | Deposit date: | 2018-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water Networks Can Determine the Affinity of Ligand Binding to Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
3QFB
 
 | | Crystal structure of the human thioredoxin reductase-thioredoxin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Thioredoxin, ... | | Authors: | Fritz-Wolf, K, Kehr, S, Stumpf, M, Rahlfs, S, Becker, K. | | Deposit date: | 2011-01-21 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human thioredoxin reductase-thioredoxin complex
Nat Commun, 2, 2011
|
|
4IZH
 
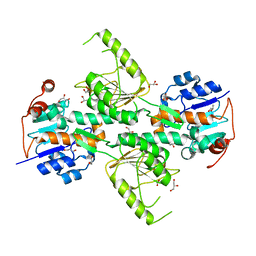 | |
4ISQ
 
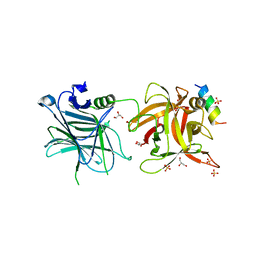 | | Binding domain of Botulinum neurotoxin DC in complex with human synaptotagmin I | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION, ... | | Authors: | Berntsson, R.P.-A, Peng, L, Svensson, L.M, Dong, M, Stenmark, P. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin DC in Complex with Its Protein Receptors Synaptotagmin I and II.
Structure, 21, 2013
|
|
3CJ7
 
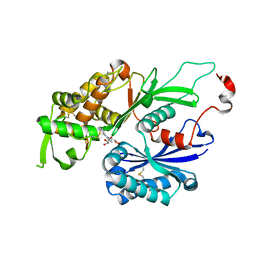 | | Structure of Rattus norvegicus NTPDase2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ectonucleoside triphosphate diphosphohydrolase 2 | | Authors: | Zebisch, M, Strater, N. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6GUR
 
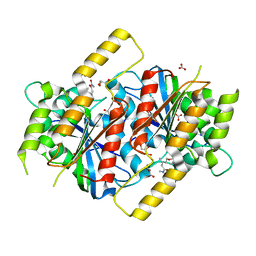 | | Siderophore hydrolase EstB from Aspergillus fumigatus in complex with TAFC | | Descriptor: | (~{Z})-5-[(1~{S},2~{S})-2-acetamido-1-oxidanyl-5-[oxidanyl(propanoyl)amino]pentoxy]-~{N},3-dimethyl-~{N}-oxidanyl-pent-2-enamide, CARBONATE ION, FE (III) ION, ... | | Authors: | Ecker, F, Haas, H, Groll, M, Huber, E.M. | | Deposit date: | 2018-06-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron Scavenging in Aspergillus Species: Structural and Biochemical Insights into Fungal Siderophore Esterases.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3QML
 
 | | The structural analysis of Sil1-Bip complex reveals the mechanism for Sil1 to function as a novel nucleotide exchange factor | | Descriptor: | 78 kDa glucose-regulated protein homolog, MAGNESIUM ION, Nucleotide exchange factor SIL1, ... | | Authors: | Yan, M, Li, J.Z, Sha, B.D. | | Deposit date: | 2011-02-04 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor.
Biochem.J., 438, 2011
|
|
3QNM
 
 | | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Matthew, M.W, Ramagopal, U.A, Toro, R, Dickey, M, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function
To be Published
|
|
3CJJ
 
 | | Crystal structure of human rage ligand-binding domain | | Descriptor: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | Authors: | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | Deposit date: | 2008-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
1HM3
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
6GF2
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
6GQB
 
 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GDP+AlF4/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
6GWP
 
 | | Crystal Structure of Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-stab) in Complex with Two Inhibitory Nanobodies (VHH-2g-42, VHH-2w-64) | | Descriptor: | Plasminogen Activator Inhibitor-1, VHH-2g-42, VHH-2w-64 | | Authors: | Sillen, M, Weeks, S.D, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2018-06-25 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.
J.Thromb.Haemost., 18, 2020
|
|
4J4Z
 
 | | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Designed serine hydrolase variant OSH55.4_H1.2, ... | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301
To be Published
|
|
