6TT0
 
 | | Crystal structure of a potent and reversible dual binding site Acetylcholinesterase chiral inhibitor | | Descriptor: | (1~{R},3~{S})-~{N}-(6,7-dimethoxy-2-oxidanylidene-chromen-3-yl)-3-[(phenylmethyl)amino]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | de la Mora, E, Mangiatordi, G.F, Belviso, B.D, Caliandro, R, Colletier, J.P, Catto, M. | | Deposit date: | 2019-12-22 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.80003023 Å) | | Cite: | Chiral Separation, X-ray Structure, and Biological Evaluation of a Potent and Reversible Dual Binding Site AChE Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
4N7M
 
 | |
3R5O
 
 | | Crystal structure of the complex of bovine lactoperoxidase with 4-allyl-2-methoxyphenol at 2.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pandey, N, Singh, A.K, Singh, R.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-03-19 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of bovine lactoperoxidase with 4-allyl-2-methoxyphenol at 2.6 A resolution
To be Published
|
|
6TT8
 
 | | Haddock model of NDM-1/morin complex | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Riviere, G, Oueslati, S, Gayral, M, Crechet, J.B, Nhiri, N, Jacquet, E, Cintrat, J.C, Giraud, F, van Heijenoort, C, Lescop, E, Pethe, S, Iorga, B.I, Naas, T, Guittet, E, Morellet, N. | | Deposit date: | 2019-12-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Characterization of the Influence of Zinc(II) Ions on the Structural and Dynamic Behavior of the New Delhi Metallo-beta-Lactamase-1 and on the Binding with Flavonols as Inhibitors.
Acs Omega, 5, 2020
|
|
4MV5
 
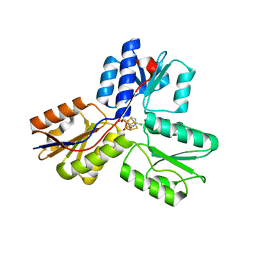 | | IspH in complex with 6-chloropyridin-3-ylmethyl diphosphate | | Descriptor: | (6-chloropyridin-3-yl)methyl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
1IUY
 
 | |
4IOV
 
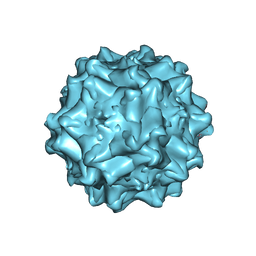 | | The structure of AAVrh32.33, a Novel Gene Delivery Vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein VP1 | | Authors: | Mikalism, K, Nam, H.-J, Van vilet, K, Vandenberghe, L.H, Mays, L.E, McKenna, R, Wilson, J, Agbandje-McKenna, M. | | Deposit date: | 2013-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of AAVrh32.33, a novel gene delivery vector.
J.Struct.Biol., 186, 2014
|
|
4HC4
 
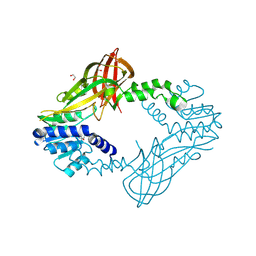 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
3ETG
 
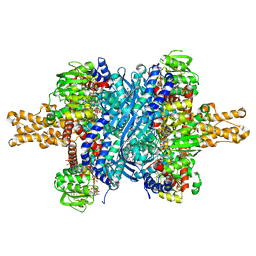 | | Glutamate dehydrogenase complexed with GW5074 | | Descriptor: | (3E)-3-[(3,5-dibromo-4-hydroxyphenyl)methylidene]-5-iodo-1,3-dihydro-2H-indol-2-one, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, M, Smith, T.J. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel Inhibitors Complexed with Glutamate Dehydrogenase: ALLOSTERIC REGULATION BY CONTROL OF PROTEIN DYNAMICS
J.Biol.Chem., 284, 2009
|
|
3R2M
 
 | | 1.8A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
4ICA
 
 | | Crystal structure of a C-terminal truncated form of the matrix subunit (p15) of Feline Immunodeficiency Virus (FIV) | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Matrix protein p15 | | Authors: | Serriere, J, Robert, X, Perez, M, Gouet, P, Guillon, C. | | Deposit date: | 2012-12-10 | | Release date: | 2013-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biophysical characterization and crystal structure of the Feline Immunodeficiency Virus p15 matrix protein.
Retrovirology, 10, 2013
|
|
3R77
 
 | | Crystal structure of the D38A mutant of isochorismatase PhzD from Pseudomonas fluorescens 2-79 in complex with 2-amino-2-desoxyisochorismate ADIC | | Descriptor: | (5S,6S)-6-amino-5-[(1-carboxyethenyl)oxy]cyclohexa-1,3-diene-1-carboxylic acid, CHLORIDE ION, Probable isochorismatase | | Authors: | Li, Q.A, Mavrodi, D.V, Thomashow, L.S, Roessle, M, Blankenfeldt, W. | | Deposit date: | 2011-03-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Binding Induces an Ammonia Channel in 2-Amino-2-desoxyisochorismate (ADIC) Synthase PhzE.
J.Biol.Chem., 286, 2011
|
|
6TQV
 
 | |
1IJI
 
 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
3R2T
 
 | | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325. | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Falugi, F, Bottomley, M, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325.
TO BE PUBLISHED
|
|
6HEH
 
 | | Structure of the catalytic domain of USP28 (insertion deleted) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 28,Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
6GK0
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH CLASS III HISTONE DEACETYLASE INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 4-~{tert}-butyl-~{N}-[[4-[5-(dimethylamino)pentanoylamino]phenyl]carbamothioyl]benzamide, ACETIC ACID, ... | | Authors: | Hakansson, M, Ladds, M.J.G.W, Walse, B, Lain, S. | | Deposit date: | 2018-05-17 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J.Biol.Chem., 295, 2020
|
|
4ICW
 
 | |
6TQX
 
 | |
3ERM
 
 | |
6HEI
 
 | |
3R7T
 
 | | Crystal Structure of Adenylosuccinate Synthetase from Campylobacter jejuni | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-23 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Adenylosuccinate Synthetase from
Campylobacter jejuni
To be Published
|
|
6HEL
 
 | | Structure of human USP25 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Gersch, M, Komander, D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Distinct USP25 and USP28 Oligomerization States Regulate Deubiquitinating Activity.
Mol.Cell, 74, 2019
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
3R45
 
 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
