2MXQ
 
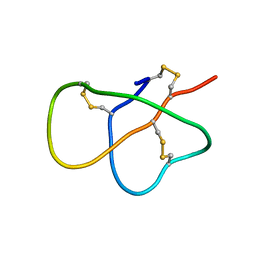 | | The solution structure of DEFA1, a highly potent antimicrobial peptide from the horse | | Descriptor: | Paneth cell-specific alpha-defensin 1 | | Authors: | Jung, S, Michalek, M, Shomali, M, Soennichsen, F.D. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional studies of the highly potent equine antimicrobial peptide DEFA1.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
5UEN
 
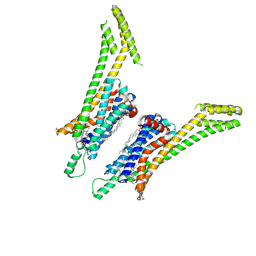 | | Crystal structure of the human adenosine A1 receptor A1AR-bRIL in complex with the covalent antagonist DU172 at 3.2A resolution | | Descriptor: | 4-{[3-(8-cyclohexyl-2,6-dioxo-1-propyl-1,2,6,7-tetrahydro-3H-purin-3-yl)propyl]carbamoyl}benzene-1-sulfonyl fluoride, Adenosine receptor A1,Soluble cytochrome b562,Adenosine receptor A1, OLEIC ACID | | Authors: | Glukhova, A, Thal, D.M, Nguyen, A.T, Vecchio, E.A, Jorg, M, Scammells, P.J, May, L.T, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity.
Cell, 168, 2017
|
|
7QD3
 
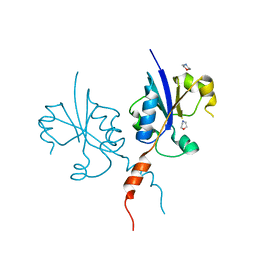 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with morpholine | | Descriptor: | Cholinephosphate cytidylyltransferase, morpholine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
7JTL
 
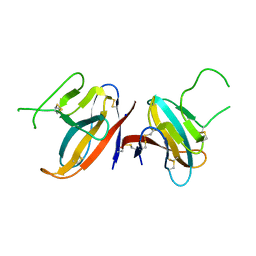 | | Structure of SARS-CoV-2 ORF8 accessory protein | | Descriptor: | ORF8 protein, SODIUM ION | | Authors: | Flower, T.G, Buffalo, C.Z, Hooy, R.M, Allaire, M, Ren, X, Hurley, J.H. | | Deposit date: | 2020-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1AUA
 
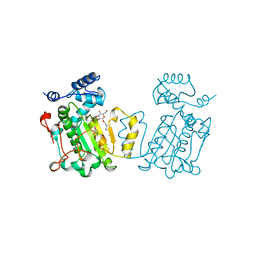 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
7QEK
 
 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-piruvic acid (IPA). | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, MAGNESIUM ION, regulator AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
6H8T
 
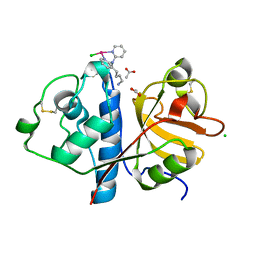 | | Crystal structure of Papain modify by achiral Ru(II)complex | | Descriptor: | ACETATE ION, CHLORIDE ION, Papain, ... | | Authors: | Cherrier, M.V, Amara, P, Talbi, B, Salmin, M, Fontecilla-Camps, J.C. | | Deposit date: | 2018-08-03 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic evidence for unexpected selective tyrosine hydroxylations in an aerated achiral Ru-papain conjugate.
Metallomics, 10, 2018
|
|
5J01
 
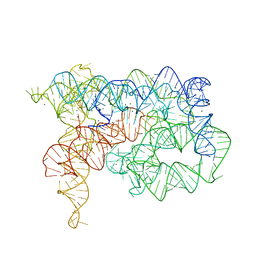 | | Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+ and MG2+. | | Descriptor: | AMMONIUM ION, MAGNESIUM ION, group II intron lariat | | Authors: | Costa, M, Walbott, H, Monachello, D, Westhof, E, Michel, F. | | Deposit date: | 2016-03-26 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structures of a group II intron lariat primed for reverse splicing.
Science, 354, 2016
|
|
7QEJ
 
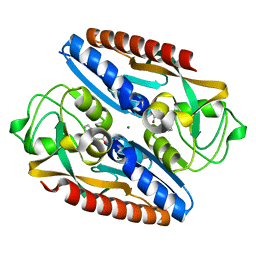 | | Structure of the ligand binding domain of the antibiotic biosynthesis regulator AdmX from the rhizobacterium Serratia plymuthica A153 bound to the auxin indole-3-acetic acid (IAA). | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MAGNESIUM ION, TRANSCRIPTIONAL REGULATOR AdmX | | Authors: | Gavira, J.A, Rico-Jimenez, M, Castellvi, A, Krell, T, Matilla, M.A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Emergence of an Auxin Sensing Domain in Plant-Associated Bacteria.
Mbio, 14, 2023
|
|
8DEW
 
 | |
5J0G
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, circular permutant P7/8 | | Descriptor: | OXIDOREDUCTASE,Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Lang, L, Logan, D, Danielsson, J, Oliveberg, M. | | Deposit date: | 2016-03-28 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tricking a Protein To Swap Strands.
J. Am. Chem. Soc., 138, 2016
|
|
5J26
 
 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
5GZS
 
 | | Structure of VC protein | | Descriptor: | ARGININE, GGDEF family protein | | Authors: | Xu, M, Wang, Y.Z, Yang, X.A, Xie, W, Jiang, T. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural studies of the periplasmic portion of the diguanylate cyclase CdgH from Vibrio cholerae.
Sci Rep, 7, 2017
|
|
7QTR
 
 | | GB1 in mammalian cells, 50 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
5J3V
 
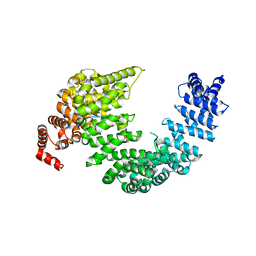 | |
7QTS
 
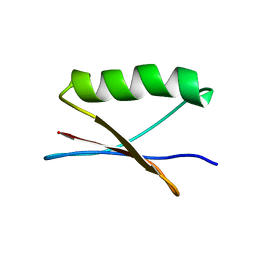 | | GB1 in mammalian cells, 10 uM | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Gerez, J.A, Prymaczok, N.C, Kadavath, H, Gosh, D, Butikofer, M, Guntert, P, Riek, R. | | Deposit date: | 2022-01-15 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in human cells by in-cell NMR and a reporter system to optimize protein delivery or transexpression.
Commun Biol, 5, 2022
|
|
5J5F
 
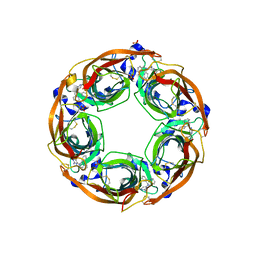 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with N4,N4-bis[(pyridin-2-yl)methyl]-6-(thiophen-3-yl)pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kaczanowska, K, Camacho Hernandez, G.A, Harel, M, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
8DEU
 
 | |
5U23
 
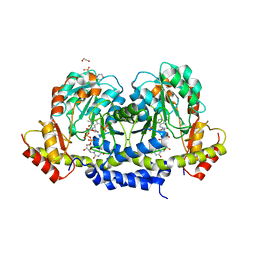 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
8DEV
 
 | |
7QUJ
 
 | | Structure of NsNEPS2, a 7S-cis-trans nepetalactone synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NsNEPS2 | | Authors: | Hernandez Lozada, N.J, Hong, B, Wood, J.C, Caputi, L, Basquin, J, Chuang, L, Kunert, M, Rodriguez Lopez, C.R, Langley, C, Zhao, D, Buell, C.R, Lichman, B.R, O'Connor, S.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biocatalytic routes to stereo-divergent iridoids.
Nat Commun, 13, 2022
|
|
2MUF
 
 | | Binding activity, structure, and immunogenicity of synthetic peptides derived from Plasmodium falciparum CelTOS and TRSP proteins | | Descriptor: | TRSP | | Authors: | Curtidor, H, Arevalo-Pinzon, G, Bermudez, A, Calderon, D, Vanegas, M, Patino, L, Patarroyo, M. | | Deposit date: | 2014-09-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Binding activity, structure, and immunogenicity of synthetic peptides derived from Plasmodium falciparum CelTOS and TRSP proteins.
Amino Acids, 43, 2012
|
|
7QJR
 
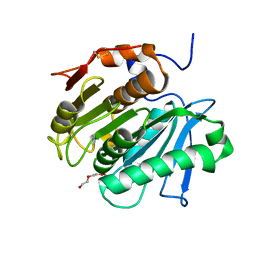 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
5GMV
 
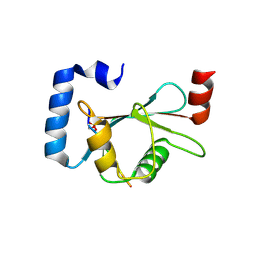 | | LC3B-FUNDC1 complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FUN14 domain-containing protein 1 | | Authors: | Lv, M, Wang, C, Li, F. | | Deposit date: | 2016-07-17 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy
Protein Cell, 8, 2017
|
|
5CKS
 
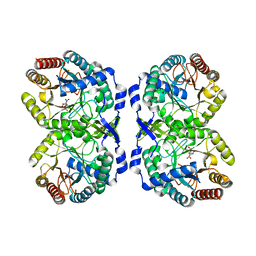 | | DAHP (3-deoxy-D-arabinoheptulosonate-7-phosphate) Synthase in complex with DAHP Oxime. | | Descriptor: | DAHP Oxime, Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive, ... | | Authors: | Berti, P, Junop, M, Balachandran, N. | | Deposit date: | 2015-07-15 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1181 Å) | | Cite: | Potent Inhibition of 3-Deoxy-d-arabinoheptulosonate-7-phosphate (DAHP) Synthase by DAHP Oxime, a Phosphate Group Mimic.
Biochemistry, 55, 2016
|
|
