5FTE
 
 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP-AlF3 and ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5IFJ
 
 | | Crystal structure of anti-gliadin 1002-1E01 Fab fragment in complex of peptide PLQPEQPFP | | Descriptor: | 1E01 Fab fragment heavy chain, 1E01 Fab fragment light chain, SULFATE ION, ... | | Authors: | Snir, O, Chen, X, Gidoni, M, du Pre, M.F, Zhao, Y, Steinsbo, O, Lundin, K.E, Yaari, G, Sollid, L.M. | | Deposit date: | 2016-02-26 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
JCI Insight, 2, 2017
|
|
7KK1
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, N84A mutant with pyruvate bound in the active site and L-lysine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-27 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A TIGHT DIMER INTERFACE N84 RESIDUE, PLAYS A CRITICAL ROLE IN THE TRANSMISSION OF THE ALLOSTERIC INHIBITION SIGNALS IN Cj.DHDPS
To Be Published
|
|
7QU1
 
 | | Machupo virus GP1 glycoprotein in complex with Fab fragment of antibody MAC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab MAC1 heavy chain, Fab MAC1 light chain, ... | | Authors: | Ng, W.M, Sahin, M, Krumm, S.A, Seow, J, Zeltina, A, Harlos, K, Paesen, G, Pinschewer, D.D, Doores, K.J, Bowden, T.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Contrasting Modes of New World Arenavirus Neutralization by Immunization-Elicited Monoclonal Antibodies.
Mbio, 13, 2022
|
|
5IG3
 
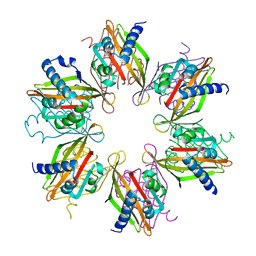 | | Crystal structure of the human CaMKII-alpha hub | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | McSpadden, E, Cao, Y.M, Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
8BZZ
 
 | |
1BTS
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | Descriptor: | BAND 3 ANION TRANSPORT PROTEIN | | Authors: | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | Deposit date: | 1994-08-03 | | Release date: | 1994-11-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
7QU2
 
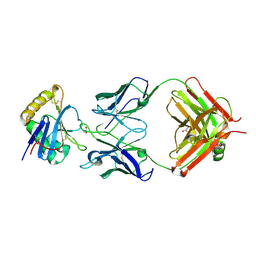 | | Junin virus GP1 glycoprotein in complex with Fab fragment of antibody JUN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab JUN1 heavy chain, ... | | Authors: | Ng, W.M, Sahin, M, Krumm, S.A, Seow, J, Zeltina, A, Harlos, K, Paesen, G, Pinschewer, D.D, Doores, K.J, Bowden, T.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contrasting Modes of New World Arenavirus Neutralization by Immunization-Elicited Monoclonal Antibodies.
Mbio, 13, 2022
|
|
5G0J
 
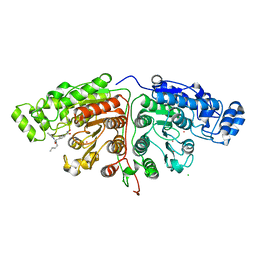 | | Crystal structure of Danio rerio HDAC6 CD1 and CD2 (linker intact) in complex with Nexturastat A | | Descriptor: | CHLORIDE ION, HDAC6, NEXTURASTAT A, ... | | Authors: | Miyake, Y, Keusch, J.J, Wang, L, Saito, M, Hess, D, Wang, X, Melancon, B.J, Helquist, P, Gut, H, Matthias, P. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into Hdac6 Tubulin Deacetylation and its Selective Inhibition
Nat.Chem.Biol., 12, 2016
|
|
8C1T
 
 | |
1BT6
 
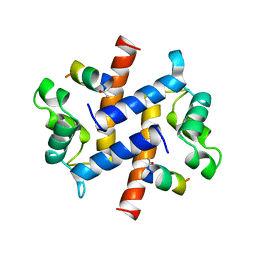 | | P11 (S100A10), LIGAND OF ANNEXIN II IN COMPLEX WITH ANNEXIN II N-TERMINUS | | Descriptor: | ANNEXIN II, S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-09-02 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
5ILD
 
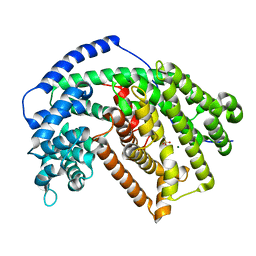 | | Tobacco 5-epi-aristolochene synthase with MES buffer molecule and Mg2+ ions | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-epi-aristolochene synthase, MAGNESIUM ION | | Authors: | Koo, H.J, Louie, G.V, Xu, Y, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-04 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Small-molecule buffer components can directly affect terpene-synthase activity by interacting with the substrate-binding site of the enzyme
To Be Published
|
|
5FIQ
 
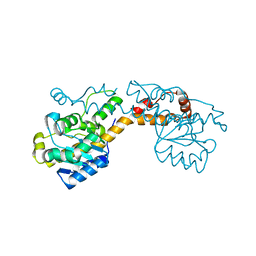 | | Exonuclease domain-containing 1 (Exd1) in the native conformation | | Descriptor: | EXD1 | | Authors: | Yang, Z, Chen, K.M, Pandey, R.R, Homolka, D, Reuter, M, Rodino Janeiro, B.K, Sachidanandam, R, Fauvarque, M.O, McCarthy, A.A, Pillai, R.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Piwi Slicing and Exd1 Drive Biogenesis of Nuclear Pirnas from Cytosolic Targets of the Mouse Pirna Pathway
Mol.Cell, 61, 2016
|
|
7QFQ
 
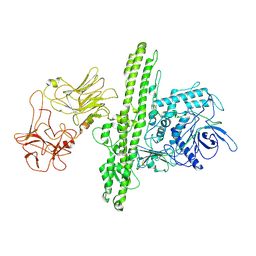 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
1BXY
 
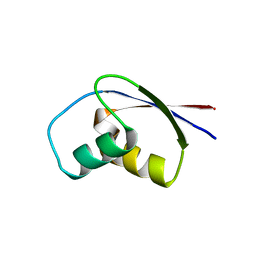 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5TVA
 
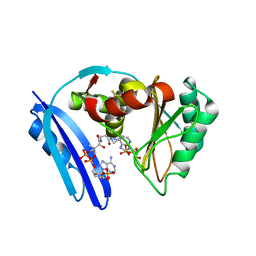 | | A. aeolicus BioW with AMP and CoA | | Descriptor: | 6-carboxyhexanoate--CoA ligase, ADENOSINE MONOPHOSPHATE, COENZYME A | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5IOB
 
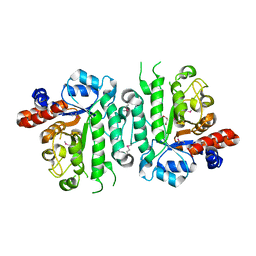 | | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase-related glycosidases, CHLORIDE ION, ... | | Authors: | Chang, C, Mack, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-08 | | Release date: | 2016-03-23 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum
To Be Published
|
|
5IIK
 
 | | Crystal structure of the post-catalytic nick complex of DNA polymerase lambda with a templating 8-oxo-dG and incorporated dC | | Descriptor: | CITRIC ACID, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*(8OG)P*TP*AP*CP*TP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
5U3Q
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 1 | | Descriptor: | 6-(2-{[([1,1'-biphenyl]-4-carbonyl)(propan-2-yl)amino]methyl}phenoxy)hexanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5IIQ
 
 | |
1BWZ
 
 | |
1C04
 
 | | IDENTIFICATION OF KNOWN PROTEIN AND RNA STRUCTURES IN A 5 A MAP OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI | | Descriptor: | 23S RRNA FRAGMENT, RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L14, ... | | Authors: | Ban, N, Nissen, P, Capel, M, Moore, P.B, Steitz, T.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit.
Nature, 400, 1999
|
|
5IJJ
 
 | |
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
5IJP
 
 | |
