6CW3
 
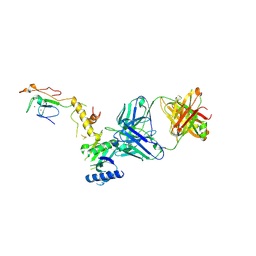 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VJE
 
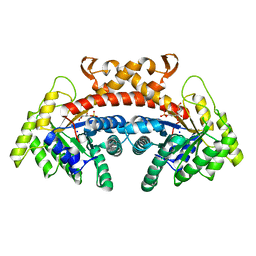 | | Class II fructose-1,6-bisphosphate aldolase of Escherichia coli with D-glucitol 1,6-bisphosphate | | Descriptor: | D-Glucitol-1,6-bisphosphate, Fructose-bisphosphate aldolase class 2, SODIUM ION | | Authors: | Coincon, M, Sygusch, J. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active site remodeling during the catalytic cycle in metal-dependent fructose-1,6-bisphosphate aldolases.
J. Biol. Chem., 293, 2018
|
|
7KEP
 
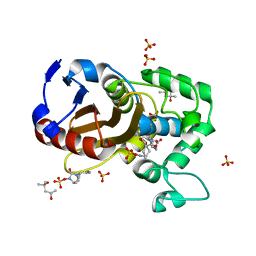 | | avibactam-CDD-1 2 minute complex | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2S,5R)-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-05-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibition of the Clostridioides difficile Class D beta-Lactamase CDD-1 by Avibactam.
Acs Infect Dis., 7, 2021
|
|
2OVJ
 
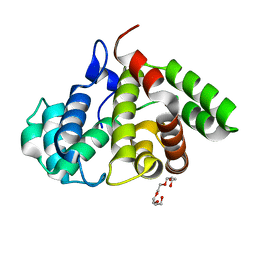 | | The crystal structure of the human Rac GTPase activating protein 1 (RACGAP1) MgcRacGAP. | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Rac GTPase-activating protein 1 | | Authors: | Shrestha, L, Papagrigoriou, E, Soundararajan, M, Elkins, J, Johansson, C, von Delft, F, Pike, A.C.W, Burgess, N, Turnbull, A, Debreczeni, J, Gorrec, F, Umeano, C, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-14 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The crystal structure of the human Rac GTPase activating protein 1 (RACGAP1) MgcRacGAP.
To be Published
|
|
5VOP
 
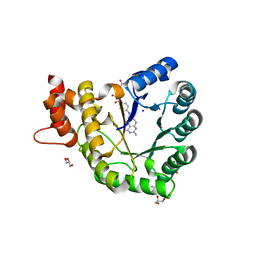 | | Methionine synthase folate-binding domain from Thermus thermophilus HB8 native | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate homocysteine S-methyltransferase, CITRATE ANION, ... | | Authors: | Koutmos, M, Yamada, K. | | Deposit date: | 2017-05-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The folate-binding module of Thermus thermophilus cobalamin-dependent methionine synthase displays a distinct variation of the classical TIM barrel: a TIM barrel with a `twist'.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5VMO
 
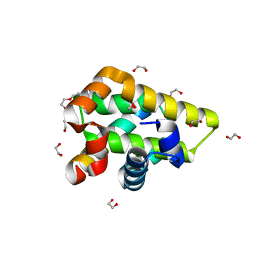 | | Crystal structure of grouper iridovirus GIV66:Bim complex | | Descriptor: | 1,2-ETHANEDIOL, Bak protein, Bcl-2 interacting mediator of cell death, ... | | Authors: | Banjara, S, Kvansakul, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.
J. Biol. Chem., 293, 2018
|
|
3CI4
 
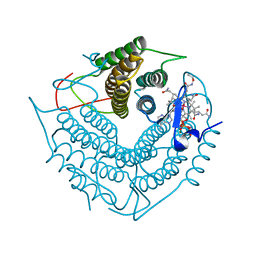 | | Structure of the PduO-type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri complexed with four-coordinate cob(II)inamide and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COB(II)INAMIDE, Cobalamin adenosyltransferase PduO-like protein, ... | | Authors: | Maurice, M.St, Mera, P.E, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a human-type corrinoid adenosyltransferase confirms that coenzyme B12 is synthesized through a four-coordinate intermediate.
Biochemistry, 47, 2008
|
|
2PCG
 
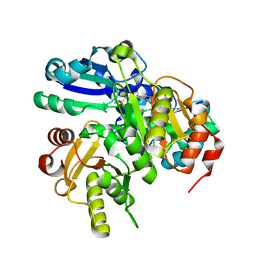 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Kageyama, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2OLN
 
 | | NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: closed form at 1.15 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PYRIDINE-2-CARBOXYLIC ACID, SODIUM ION, ... | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-01-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | NikD, an unusual amino acid oxidase essential for nikkomycin biosynthesis: structures of closed and open forms at 1.15 and 1.90 A resolution
Structure, 15, 2007
|
|
3AJ1
 
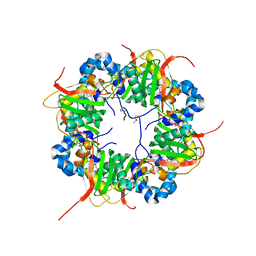 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ABZ
 
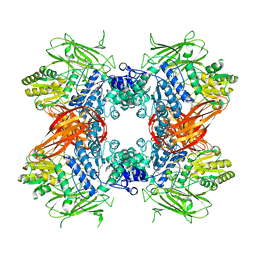 | | Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus | | Descriptor: | Beta-glucosidase I, GLYCEROL | | Authors: | Yoshida, E, Hidaka, M, Fushinobu, S, Katayama, T, Kumagai, H. | | Deposit date: | 2009-12-25 | | Release date: | 2010-08-11 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus.
Biochem.J., 431, 2010
|
|
5VY3
 
 | |
5TAC
 
 | |
5TDW
 
 | | Set3 PHD finger in complex with histone H3K4me3 | | Descriptor: | SET domain-containing protein 3, SODIUM ION, ZINC ION, ... | | Authors: | Andrews, F.H, Ali, M, Kutateladze, T.G. | | Deposit date: | 2016-09-19 | | Release date: | 2016-10-19 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insight into Recognition of Methylated Histone H3K4 by Set3.
J. Mol. Biol., 429, 2017
|
|
5T6K
 
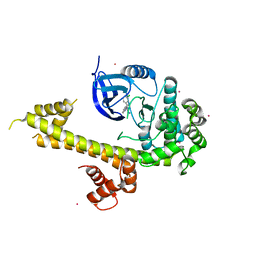 | | Crystal Structure of TgCDPK1 From Toxoplasma Gondii complexed with GW780159X | | Descriptor: | 4-(3-chlorophenyl)-5-(1,5-naphthyridin-2-yl)-1,3-thiazol-2-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Osman, K.T, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CDPK1 from toxoplasma gondii complexed with SGC-1-19
to be published
|
|
6CT8
 
 | |
5T72
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5T7J
 
 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1L6E
 
 | | Solution structure of the docking and dimerization domain of protein kinase A II-alpha (RIIalpha D/D). Alternatively called the N-terminal dimerization domain of the regulatory subunit of protein kinase A. | | Descriptor: | cAMP-dependent protein kinase Type II-alpha regulatory chain | | Authors: | Morikis, D, Roy, M, Newlon, M.G, Scott, J.D, Jennings, P.A. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Electrostatic properties of the structure of the docking and dimerization domain of protein kinase A IIalpha
Eur.J.Biochem., 269, 2002
|
|
7LKJ
 
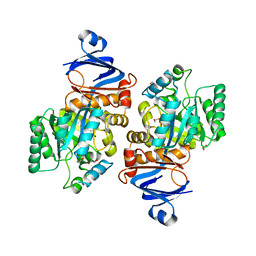 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) | | Descriptor: | 1,2-ETHANEDIOL, Aminofutalosine deaminase, FE (III) ION | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
7LKK
 
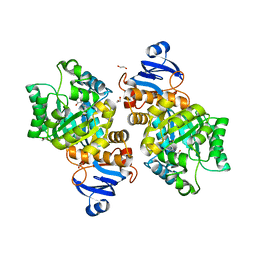 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) in complex with Methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 1,2-ETHANEDIOL, Aminofutalosine deaminase, ... | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
6BYR
 
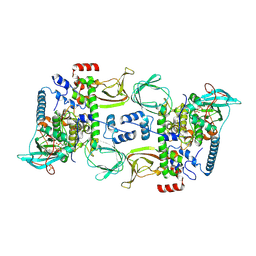 | | Structures of the PKA RI alpha holoenzyme with the FLHCC driver J-PKAc alpha or native PKAc alpha | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha chimera, MAGNESIUM ION, ... | | Authors: | Cao, B, Lu, T.W, Martinez Fiesco, J.A, Tomasini, M, Fan, L, Simon, S.M, Taylor, S.S, Zhang, P. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.661 Å) | | Cite: | Structures of the PKA RI alpha Holoenzyme with the FLHCC Driver J-PKAc alpha or Wild-Type PKAc alpha.
Structure, 27, 2019
|
|
5TCQ
 
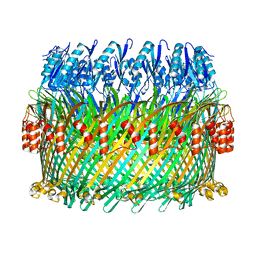 | | Near-atomic resolution cryo-EM structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG | | Descriptor: | Protein InvG | | Authors: | Worrall, L.J, Hong, C, Vuckovic, M, Bergeron, J.R.C, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2016-09-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body.
Nature, 540, 2016
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
2P9Z
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
