7UVN
 
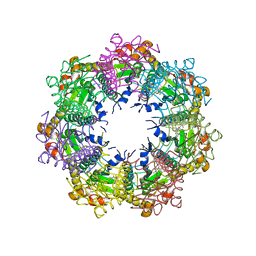 | | Crystal structure of human ClpP protease in complex with TR-57 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-1-methyl-2,4-dioxo-1,3,4,5,7,8-hexahydropyrido[4,3-d]pyrimidin-6(2H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UVU
 
 | | Crystal structure of human ClpP protease in complex with TR-107 | | Descriptor: | 3-({3-[(4-chlorophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
7UW0
 
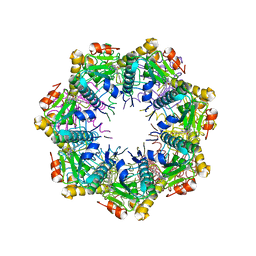 | | Crystal structure of human ClpP protease in complex with TR-133 | | Descriptor: | 3-({3-[(4-bromophenyl)methyl]-4-oxo-3,5,7,8-tetrahydropyrido[4,3-d]pyrimidin-6(4H)-yl}methyl)benzonitrile, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Mabanglo, M.F, Houry, W.A. | | Deposit date: | 2022-05-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome.
Structure, 31, 2023
|
|
8IFU
 
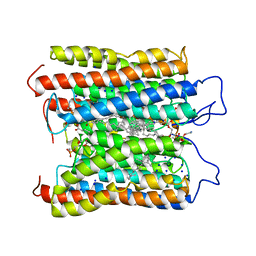 | | HcCCR in NaCl | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, HcCCR, RETINAL, ... | | Authors: | Zhang, M.F. | | Deposit date: | 2023-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structure of HcCCR
To Be Published
|
|
7VLJ
 
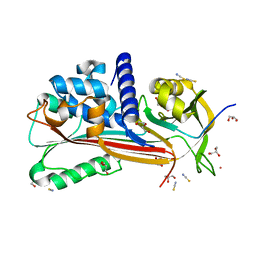 | | Crystal structure of Entamoeba histolytica serine protease inhibitor, Histopin, in the cleaved conformation | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ali, M.F, Devi, S, Gourinath, S. | | Deposit date: | 2021-10-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Entamoeba histolytica serine protease inhibitor, Histopin, in the cleaved conformation
To Be Published
|
|
8IWC
 
 | | Crystal structure of Q9PR55 at pH 6.0 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWA
 
 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWB
 
 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IO5
 
 | | Herg1a-herg1b closed state 2 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
8IOB
 
 | | Herg1a-herg1b closed state 1 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Herg1a-herg1b closed state 1
To Be Published
|
|
5TZU
 
 | | Crystal structure of human CD47 ECD bound to Fab of B6H12.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
5TZT
 
 | | Crystal structure of human CD47 ECD bound to Fab of C47B161 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy Chain of Fab C47B161, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
4UWN
 
 | | Lysozyme soaked with a ruthenium based CORM with a methione oxide ligand (complex 6b) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
5TZ2
 
 | | Crystal structure of human CD47 ECD bound to Fab of C47B222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C47B222 Fab Heavy Chain, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
6IBO
 
 | | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, ALA-VAL-SER-ARG-ALA, PHOSPHATE ION, ... | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5UFU
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1,4:3,6-dianhydro-2-O-(6-chloro-5-{4-[1-(hydroxymethyl)cyclopropyl]phenyl}-1H-benzimidazol-2-yl)-D-mannitol, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2017-01-05 | | Release date: | 2017-05-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Activation of Skeletal Muscle AMPK Promotes Glucose Disposal and Glucose Lowering in Non-human Primates and Mice.
Cell Metab., 25, 2017
|
|
8W68
 
 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
4WPG
 
 | | Group A Streptococcus GacA is an essential dTDP-4-dehydrorhamnose reductase (RmlD) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, ... | | Authors: | van der Beek, S.L, Le Breton, Y, Ferenbach, A.T, van Aalten, D.M.F, Navratilova, I, McIver, K, van Sorge, N.M, Dorfmueller, H.C. | | Deposit date: | 2014-10-18 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | GacA is essential for Group A Streptococcus and defines a new class of monomeric dTDP-4-dehydrorhamnose reductases (RmlD).
Mol.Microbiol., 98, 2015
|
|
4WN4
 
 | | Crystal structure of designed cPPR-polyA protein | | Descriptor: | Pentatricopeptide repeat protein | | Authors: | Coquille, S.C, Filipovska, A, Chia, T.S, Rajappa, L, Lingford, J.P, Razif, M.F.M, Thore, S, Rackham, O. | | Deposit date: | 2014-10-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | An artificial PPR scaffold for programmable RNA recognition.
Nat Commun, 5, 2014
|
|
5UU5
 
 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UZ9
 
 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
6I9F
 
 | | Solution structure of As-p18 reveals that nematode fatty acid binding proteins exhibit unusual structural features | | Descriptor: | Fatty acid-binding protein homolog, OLEIC ACID | | Authors: | Ibanez Shimabukuro, M, Rey Burusco, M.F, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | As-p18, an extracellular fatty acid binding protein of nematodes, exhibits unusual structural features.
Biosci.Rep., 2019
|
|
4WF1
 
 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8W6A
 
 | |
