5DO2
 
 | | Complex structure of MERS-RBD bound with 4C2 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C2 heavy chain, 4C2 light chain, ... | | Authors: | Li, Y, Wan, Y, Liu, P, Zhao, J, Lu, G, Qi, J, Wang, Q, Lu, X, Wu, Y, Liu, W, Yuen, K.Y, Perlman, S, Gao, G.F, Yan, J. | | Deposit date: | 2015-09-10 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein.
Cell Res., 25, 2015
|
|
4X57
 
 | |
6H9J
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (229-255) | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Leissing, T.M, Clifton, I.J, Saward, B.G, Lu, X, Hopkinson, R.J, Schofield, C.J. | | Deposit date: | 2018-08-04 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (229-255)
To Be Published
|
|
6HC8
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPA1 (313-339) | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Leissing, T.M, Clifton, I.J, Saward, B.G, Lu, X, Hopkinson, R.J, Schofield, C.J. | | Deposit date: | 2018-08-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPA1 (313-339)
To Be Published
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
8OZD
 
 | | cryoEM structure of SPARTA complex dimer-3 | | Descriptor: | DNA (16-MER), Piwi domain-containing protein, RNA (18-MER), ... | | Authors: | Ekundayo, B, Ni, D.C, Lu, X.H, Stahlberg, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
8OZC
 
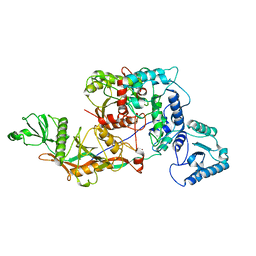 | | cryoEM structure of SPARTA complex heterodimer apo | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Ekundayo, B, Ni, D.C, Lu, X.H, Stahlberg, H. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation mechanism of a short argonaute-TIR prokaryotic immune system.
Sci Adv, 9, 2023
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
1DRS
 
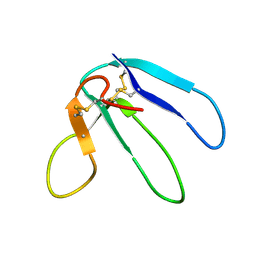 | | THREE-DIMENSIONAL STRUCTURE OF THE RGD-CONTAINING NEUROTOXIN HOMOLOGUE, DENDROASPIN | | Descriptor: | DENDROASPIN | | Authors: | Sutcliffe, M.J, Jaseja, M, Hyde, E.I, Lu, X, Williams, J.A. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the RGD-containing neurotoxin homologue dendroaspin.
Nat.Struct.Biol., 1, 1994
|
|
2VGE
 
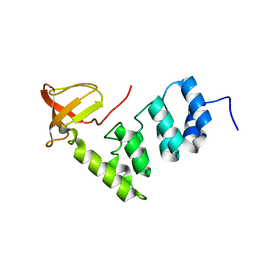 | | Crystal structure of the C-terminal region of human iASPP | | Descriptor: | RELA-ASSOCIATED INHIBITOR | | Authors: | Robinson, R.A, Lu, X, Jones, E.Y, Siebold, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and Structural Studies of Aspp Proteins Reveal Differential Binding to P53, P63 and P73
Structure, 16, 2008
|
|
4RC6
 
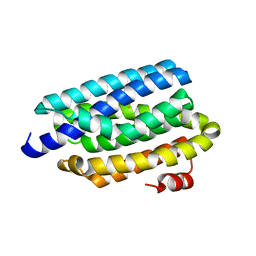 | | Crystal structure of cyanobacterial aldehyde-deformylating oxygenase 122F mutant | | Descriptor: | Aldehyde decarbonylase, FE (II) ION | | Authors: | Jia, C.J, Li, M, Li, J.J, Zhang, J.J, Zhang, H.M, Cao, P, Pan, X.W, Lu, X.F, Chang, W.R. | | Deposit date: | 2014-09-14 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases.
Protein Cell, 6, 2015
|
|
2AAF
 
 | |
4Y25
 
 | | Bacterial polysaccharide outer membrane secretin | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Wang, Y, AndolePannuri, A, Ni, D, Zhou, H, Cao, X, Lu, X, Romeo, T, Huang, Y. | | Deposit date: | 2015-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structural Basis for Translocation of a Biofilm-supporting Exopolysaccharide across the Bacterial Outer Membrane
J.Biol.Chem., 291, 2016
|
|
2LKB
 
 | | Evolutionary diversification of Mesobuthus alpha-scorpion toxins affecting sodium channels | | Descriptor: | Neurotoxin MeuNaTx-5 | | Authors: | Zhu, S, Peigneur, S, Gao, B, Lu, X, Cao, C, Tytgat, J. | | Deposit date: | 2011-10-09 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Evolutionary diversification of Mesobuthus {alpha}-scorpion toxins affecting sodium channels
Mol.Cell Proteomics, 2011
|
|
5JYY
 
 | | Structure-based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-resistant Influenza Viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-[(2-methoxyethyl)carbamoyl]-D-glycero-D-galacto-non-2-enon ic acid, CALCIUM ION, ... | | Authors: | Fu, L, Wu, Y, Bi, Y, Zhang, S, Lv, X, Qi, J, Li, Y, Lu, X, Yan, J, Gao, G.F, Li, X. | | Deposit date: | 2016-05-15 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-Resistant Influenza Viruses
J.Med.Chem., 59, 2016
|
|
7XQ5
 
 | | Crystal structure of ScIno2p-ScIno4p bound promoter DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*TP*CP*AP*CP*AP*TP*GP*CP*AP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*AP*TP*GP*TP*GP*AP*AP*AP*AP*T)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Khan, M.H, Lu, X. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA.
Int J Mol Sci, 23, 2022
|
|
3DLP
 
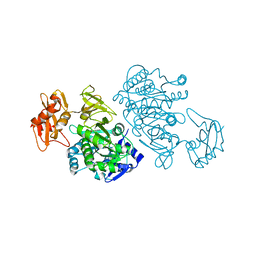 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, Mutant D402P, bound to 4CB | | Descriptor: | 4-CHLORO-BENZOIC ACID, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Cao, J, Reger, A.S, Lu, X, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2008-06-28 | | Release date: | 2009-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase
Biochemistry, 48, 2009
|
|
8GYD
 
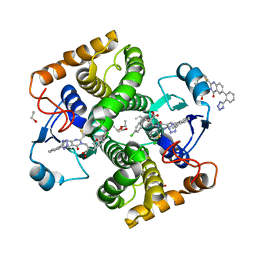 | | Structure of Schistosoma japonicum Glutathione S-transferase bound with the ligand complex of 16 | | Descriptor: | (2R)-2-[[2-(5-chloranylthiophen-2-yl)-4-oxidanylidene-6-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]quinazolin-3-yl]methyl]-3-(4-chlorophenyl)propanoic acid, ETHANOL, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Wen, X, Jin, R, Hu, H, Zhu, J, Song, W, Lu, X. | | Deposit date: | 2022-09-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery, SAR Study of GST Inhibitors from a Novel Quinazolin-4(1 H )-one Focused DNA-Encoded Library.
J.Med.Chem., 66, 2023
|
|
1FBZ
 
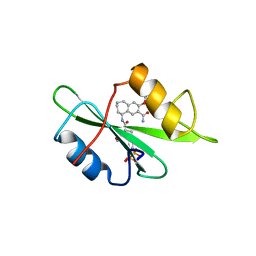 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
8FTQ
 
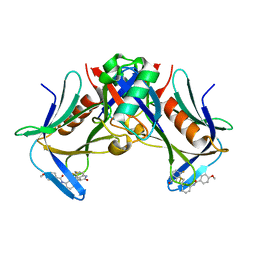 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
6MEZ
 
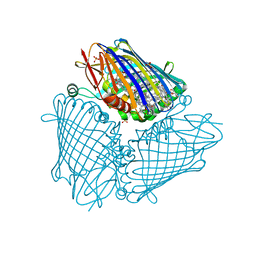 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | Authors: | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
1T5H
 
 | |
1T5D
 
 | |
1BL4
 
 | | FKBP MUTANT F36V COMPLEXED WITH REMODELED SYNTHETIC LIGAND | | Descriptor: | PROTEIN (FK506 BINDING PROTEIN), {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Hatada, M.H, Clackson, T, Yang, W, Rozamus, L.W, Amara, J, Rollins, C.T, Stevenson, L.F, Magari, S.R, Wood, S.A, Courage, N.L, Lu, X, Cerasoli Junior, F, Gilman, M, Holt, D. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6KJF
 
 | | lovastatin esterase PcEST in complex with simvastatin | | Descriptor: | Pc15g00720 protein, Simvastatin acid, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Liang, Y.J, Lu, X.F. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the catalytic mechanism of lovastatin hydrolase
J.Biol.Chem., 2019
|
|
