8SR9
 
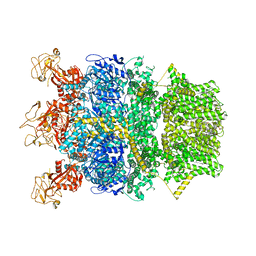 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium | | Descriptor: | CHOLESTEROL, MAGNESIUM ION, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SRA
 
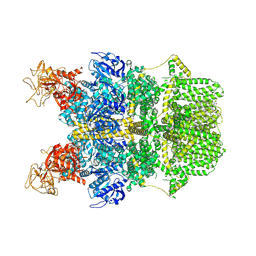 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Calcium | | Descriptor: | CALCIUM ION, CHOLESTEROL, TRPM2 chanzyme | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution.
Nat.Struct.Mol.Biol., 2024
|
|
8SRD
 
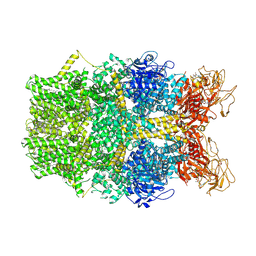 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRI
 
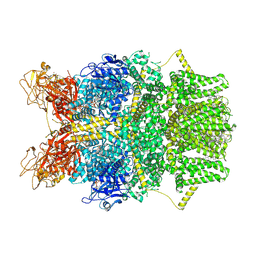 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRG
 
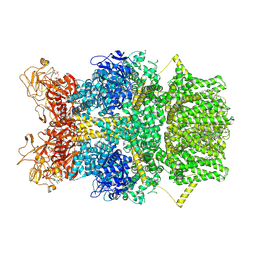 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, Adenosine monophosphate, and Ribose-5-phosphate | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRK
 
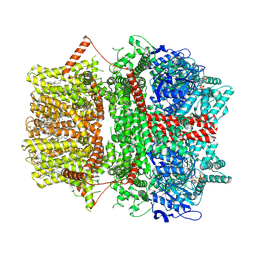 | | Cryo-EM structure of TRPM2 chanzyme (without NUDT9-H domain) in the presence of Ca and ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRH
 
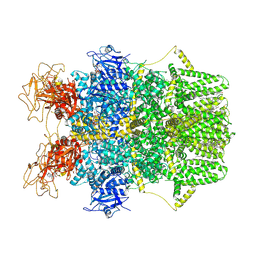 | | Cryo-EM structure of TRPM2 chanzyme (E1114A) in the presence of Magnesium and ADP-ribose, open state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
8SRF
 
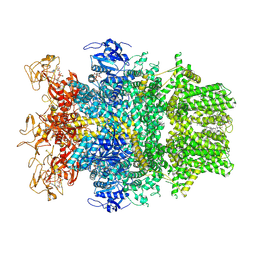 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium, ADP-ribose, Adenosine monophosphate, and Ribose-5-phosphate, closed state | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, ADENOSINE MONOPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
2C9A
 
 | | Crystal structure of the MAM-Ig module of receptor protein tyrosine phosphatase mu | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE MU, ... | | Authors: | Aricescu, A.R, Hon, W.C, Siebold, C, Lu, W, Van Der Merwe, P.A, Jones, E.Y. | | Deposit date: | 2005-12-09 | | Release date: | 2006-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Analysis of Receptor Protein Tyrosine Phosphatase Mu-Mediated Cell Adhesion.
Embo J., 25, 2006
|
|
8SRE
 
 | | Cryo-EM structure of TRPM2 chanzyme in the presence of Magnesium and ADP-ribose, closed state | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Huang, Y, Kumar, S, Lu, W, Du, J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Coupling enzymatic activity and gating in an ancient TRPM chanzyme and its molecular evolution
To Be Published
|
|
2YD3
 
 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Sigma | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE S, SODIUM ION | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD5
 
 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase LAR | | Descriptor: | RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE F | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
2YD1
 
 | | Crystal structure of the N-terminal Ig1-2 module of Drosophila Receptor Protein Tyrosine Phosphatase DLAR | | Descriptor: | GLYCINE, TYROSINE-PROTEIN PHOSPHATASE LAR | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
1ONJ
 
 | | Crystal structure of Atratoxin-b from Chinese cobra venom of Naja atra | | Descriptor: | Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Tu, X, Pan, G, Xu, C, Fan, R, Lu, W, Deng, W, Rao, P, Teng, M, Niu, L. | | Deposit date: | 2003-02-28 | | Release date: | 2004-02-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Purification, N-terminal sequencing, crystallization and preliminary structural determination of atratoxin-b, a short-chain alpha-neurotoxin from Naja atra venom.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2YD2
 
 | | Crystal structure of the N-terminal Ig1-2 module of Human Receptor Protein Tyrosine Phosphatase Sigma | | Descriptor: | CHLORIDE ION, IODIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE S | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
4TLL
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 1 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
4RSY
 
 | | Crystal structures of the Human leukotriene A4 Hydrolase complex with a potential inhibitor H7 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, ACETIC ACID, IMIDAZOLE, ... | | Authors: | Ouyang, P, Lu, W, Cui, K, Huang, J. | | Deposit date: | 2014-11-12 | | Release date: | 2015-12-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of Human Leukotriene A4 Hydrolase in complex with inhibitor H1
To be Published
|
|
4RVB
 
 | | Crystal Structure Analysis of the Human Leukotriene A4 Hydrolase | | Descriptor: | ACETATE ION, ACETIC ACID, GLYCEROL, ... | | Authors: | Ouyang, P, Lu, W, Cui, K, Huang, J. | | Deposit date: | 2014-11-25 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of the Human Leukotriene A4 Hydrolase
To be Published
|
|
4TLM
 
 | | Crystal structure of GluN1/GluN2B NMDA receptor, structure 2 | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, ... | | Authors: | Gouaux, E, Lee, C.-H, Lu, W. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | NMDA receptor structures reveal subunit arrangement and pore architecture.
Nature, 511, 2014
|
|
5DG5
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
4YLW
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wu, D, Ouyang, P, Lu, W, Huang, J. | | Deposit date: | 2015-03-06 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with No.33 compound
To Be Published
|
|
4ZL1
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18X at 1.86 A resolution | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Huang, J, Wu, D, Ouyang, P, Lu, W, Pu, J. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18XYW at 1.86 A resolution
To Be Published
|
|
2KHT
 
 | | NMR Structure of human alpha defensin HNP-1 | | Descriptor: | Neutrophil defensin 1 | | Authors: | Zhang, Y, Li, S, Doherty, T.F, Lubkowski, J, Lu, W, Li, J, Barinka, C, Hong, M. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR.
J.Mol.Biol., 397, 2010
|
|
2YGO
 
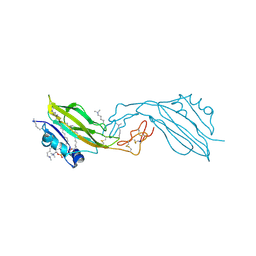 | | WIF domain-EGF-like domain 1 of human Wnt inhibitory factor 1 in complex with 1,2-dipalmitoylphosphatidylcholine | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Malinauskas, T, Aricescu, A.R, Lu, W, Siebold, C, Jones, E.Y. | | Deposit date: | 2011-04-19 | | Release date: | 2011-07-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Modular Mechanism of Wnt Signaling Inhibition by Wnt Inhibitory Factor 1
Nat.Struct.Mol.Biol., 18, 2011
|
|
3LNJ
 
 | |
