1CU2
 
 | | T4 LYSOZYME MUTANT L84M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CUQ
 
 | | T4 LYSOZYME MUTANT V103M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV0
 
 | | T4 LYSOZYME MUTANT F104M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CU5
 
 | | T4 LYSOZYME MUTANT L91M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CU3
 
 | | T4 LYSOZYME MUTANT V87M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CUP
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
5V9L
 
 | |
1FI7
 
 | | Solution structure of the imidazole complex of cytochrome C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
1FI9
 
 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, IMIDAZOLE | | Authors: | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
5V71
 
 | | KRAS G12C in bound to quinazoline based switch II pocket (SWIIP) binder | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry.
Structure, 25, 2017
|
|
3RBW
 
 | | Crystal structure of Spire KIND domain | | Descriptor: | Protein spire homolog 1 | | Authors: | Vizcarra, C.L, Kreutz, B, Rodal, A.A, Toms, A.V, Lu, J, Zheng, W, Quinlan, M.E, Eck, M.J. | | Deposit date: | 2011-03-30 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Spire KIND domain and insights into its interaction with Fmn-family formins
Proc.Natl.Acad.Sci.USA, 2011
|
|
5V9O
 
 | | KRAS G12C inhibitor | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Westover, K, Lu, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C.
Cell Chem Biol, 24, 2017
|
|
2JSZ
 
 | | Solution structure of Tpx in the reduced state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J, Yang, F. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2PVJ
 
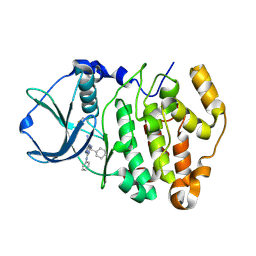 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(CYCLOHEXYLMETHYLAMINO)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PVL
 
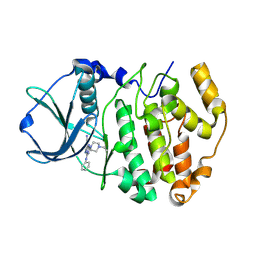 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 2-(4-ETHYLPIPERAZIN-1-YL)-4-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4PM4
 
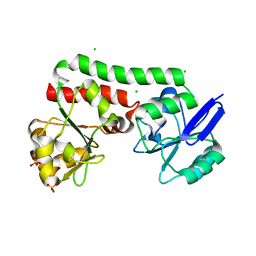 | | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv | | Descriptor: | CHLORIDE ION, Iron complex transporter substrate-binding protein, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, Tran, N, Kuo, E, Lu, J, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-05-20 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
3PKD
 
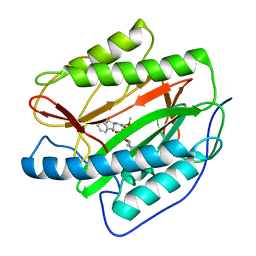 | | M. tuberculosis MetAP with bengamide analog Y10, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2,3-dihydro-1H-inden-2-yl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives.
Chemmedchem, 6, 2011
|
|
6CEW
 
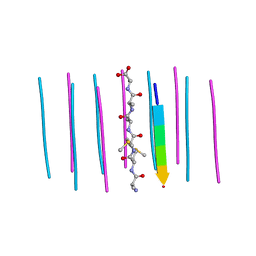 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3PKE
 
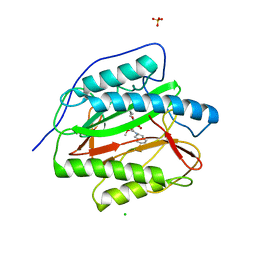 | | M. tuberculosis MetAP with bengamide analog Y10, in Ni form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2,3-dihydro-1H-inden-2-yl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives.
Chemmedchem, 6, 2011
|
|
3PKB
 
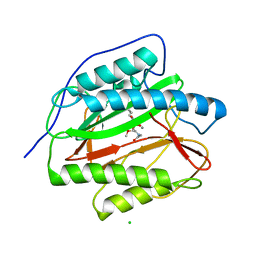 | | M. tuberculosis MetAP with bengamide analog Y16, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2-azanyl-2-oxidanylidene-ethyl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives.
Chemmedchem, 6, 2011
|
|
3PKA
 
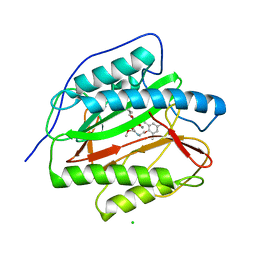 | | M. tuberculosis MetAP with bengamide analog Y02, in Mn form | | Descriptor: | (2R,3R,4S,5R,6E)-3,4,5-trihydroxy-2-methoxy-8,8-dimethyl-N-[2-(2,4,6-trimethylphenoxy)ethyl]non-6-enamide, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives.
Chemmedchem, 6, 2011
|
|
3PKC
 
 | | M. tuberculosis MetAP with bengamide analog Y08, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-[[(3S)-1-cyclopropylcarbonylpiperidin-3-yl]methyl]-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives.
Chemmedchem, 6, 2011
|
|
2KC5
 
 | |
4I0X
 
 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | Descriptor: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | Authors: | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-11-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
