6QLU
 
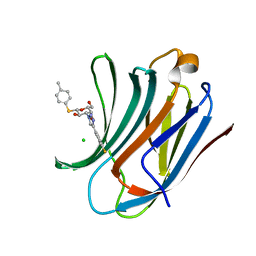 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-8 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(4-fluoranyl-3-methyl-phenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
6Y4C
 
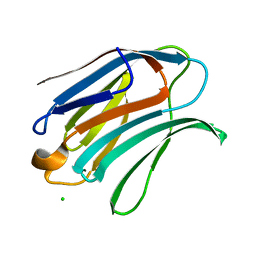 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using an XtalTool support | | Descriptor: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shilova, A, Hakansson, M, Welin, M, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
6Y78
 
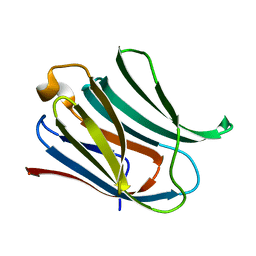 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using a silicon nitride membrane support | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hakansson, M, Welin, M, Shilova, A, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
6QLN
 
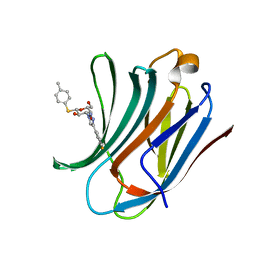 | | Galectin-3C in complex with fluoroaryl triazole monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
2BV3
 
 | | Crystal structure of a mutant elongation factor G trapped with a GTP analogue | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Mutant Elongation Factor G Trapped with a GTP Analogue.
FEBS Lett., 579, 2005
|
|
5M8E
 
 | | Crystal structure of a GH43 arabonofuranosidase from Weissella sp. strain 142 | | Descriptor: | Alpha-N-arabinofuranosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Linares-Pasten, J, Logan, D.T, Nordberg Karlsson, E. | | Deposit date: | 2016-10-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures and functional studies of two GH43 arabinofuranosidases from Weissella sp. strain 142 and Lactobacillus brevis.
FEBS J., 284, 2017
|
|
5OLK
 
 | | Crystal structure of the ATP-cone-containing NrdB from Leeuwenhoekiella blandensis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Hasan, M, Grinberg, A.R, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit.
Elife, 7, 2018
|
|
8PFF
 
 | | Galectin-3C in complex with a triazolesulfone derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-(phenylsulfonyl)-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8P23
 
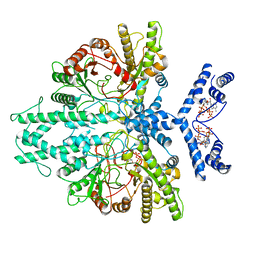 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/CTP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
Elife, 2023
|
|
8P27
 
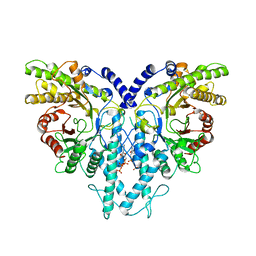 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dATP-bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductases: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
Elife, 2023
|
|
8P2C
 
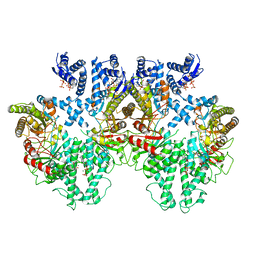 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric state produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P2S
 
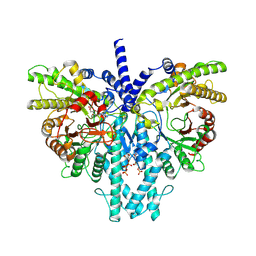 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/dTTP/GTP-bound state | | Descriptor: | Anaerobic ribonucleoside-triphosphate reductase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P2D
 
 | | Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P39
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-17 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To be published
|
|
6Q4S
 
 | | Crystal structure of a-eudesmol synthase | | Descriptor: | CHLORIDE ION, Pentalenene synthase | | Authors: | Correia Cordeiro, R.S, Hakansson, M, Logan, D.T, Kourist, R. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of three novel sesquiterpene synthases from Streptomyces chartreusis NRRL 3882 and crystal structure of an alpha-eudesmol synthase.
J.Biotechnol., 297, 2019
|
|
3KET
 
 | | Crystal structure of a Rex-family transcriptional regulatory protein from Streptococcus agalactiae bound to a palindromic operator | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*TP*CP*AP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Thiyagarajan, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2009-10-26 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | NAD+ pool depletion as a signal for the Rex regulon involved in Streptococcus agalactiae virulence.
Plos Pathog., 17, 2021
|
|
8P28
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric, dATP-bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To Be Published
|
|
3O0Q
 
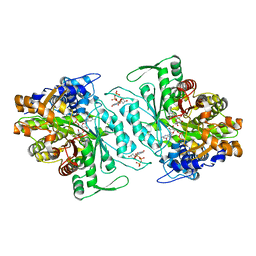 | | Thermotoga maritima Ribonucleotide Reductase, NrdJ, in complex with dTTP, GDP and Adenosine | | Descriptor: | ADENOSINE, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Logan, D.T, Nordlund, P. | | Deposit date: | 2010-07-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Adenosylcobalamin Activation in AdoCbl-Dependent Ribonucleotide Reductases.
Acs Chem.Biol., 5, 2010
|
|
3O0O
 
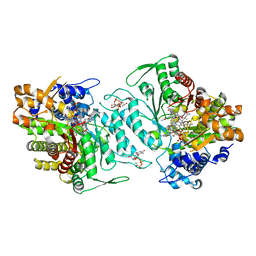 | | Thermotoga maritima Ribonucleotide Reductase, NrdJ, in complex with dTTP, GDP and Adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Larsson, K.-M, Logan, D.T, Nordlund, P. | | Deposit date: | 2010-07-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Adenosylcobalamin Activation in AdoCbl-Dependent Ribonucleotide Reductases.
Acs Chem.Biol., 5, 2010
|
|
5IUQ
 
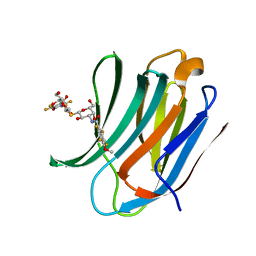 | | Galectin-3c in complex with Bisamido-thiogalactoside derivative 4 | | Descriptor: | 3-deoxy-3-[(2,3,5,6-tetrafluoro-4-methoxybenzene-1-carbonyl)amino]-beta-D-galactopyranosyl 3-deoxy-3-[(2,3,5,6-tetrafluoro-4-methoxybenzene-1-carbonyl)amino]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Noresson, A.-L, Aurelius, O, Oberg, C.T, Engstrom, O, Sundin, A.P, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2016-03-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Controlling protein:ligand complex conformation through tuning of arginine-arene interactions: Synthetic and structural studies with 3-benzamido-2-sulfo-galactosides as galectin-3 ligands
To Be Published
|
|
6QLT
 
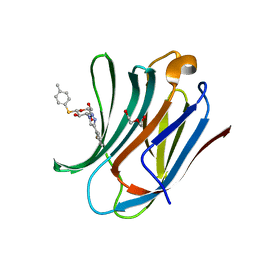 | | Galectin-3C in complex with fluoroaryltriazole monothiogalactoside derivative-7 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, DI(HYDROXYETHYL)ETHER, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-02-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Energetics of Ligand-Fluorine Interactions with Galectin-3 Backbone and Side-Chain Amides: Insight into Solvation Effects and Multipolar Interactions.
Chemmedchem, 14, 2019
|
|
5ODY
 
 | | Galectin-3C in complex with dithiogalactoside derivative | | Descriptor: | 5,6-bis(fluoranyl)-3-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]chromen-2-one, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Systematic Tuning of Fluoro-galectin-3 Interactions Provides Thiodigalactoside Derivatives with Single-Digit nM Affinity and High Selectivity.
J. Med. Chem., 61, 2018
|
|
3O0N
 
 | | Thermotoga maritima Ribonucleotide Reductase, NrdJ, in complex with dTTP and Adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, CHLORIDE ION, COBALAMIN, ... | | Authors: | Larsson, K.-M, Logan, D.T, Nordlund, P. | | Deposit date: | 2010-07-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Adenosylcobalamin Activation in AdoCbl-Dependent Ribonucleotide Reductases.
Acs Chem.Biol., 5, 2010
|
|
8PF9
 
 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[oxidanyl(phenyl)-$l^{3}-sulfanyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
6QGE
 
 | | Galectin-3C in complex with a pair of enantiomeric ligands: S enantiomer | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-[(2~{S})-3-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-2-oxidanyl-propyl]sulfanyl-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | Authors: | Manzoni, F, Verteramo, M.L, Oksanen, E, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-01-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Interplay between Conformational Entropy and Solvation Entropy in Protein-Ligand Binding.
J. Am. Chem. Soc., 141, 2019
|
|
