3TG2
 
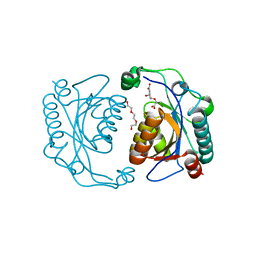 | | Crystal structure of the ISC domain of VibB in complex with isochorismate | | 分子名称: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, TRIETHYLENE GLYCOL, Vibriobactin-specific isochorismatase | | 著者 | Liu, S, Zhang, C, Niu, B, Li, N, Liu, X, Liu, M, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | 登録日 | 2011-08-17 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structural insight into the ISC domain of VibB from Vibrio cholerae at atomic resolution: a snapshot just before the enzymatic reaction
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4RM5
 
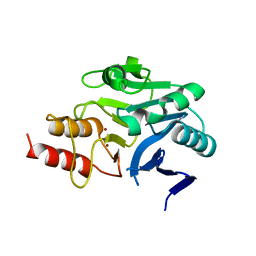 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | 分子名称: | Beta-lactamase NDM-1, ZINC ION | | 著者 | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | 登録日 | 2014-10-19 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
8WYE
 
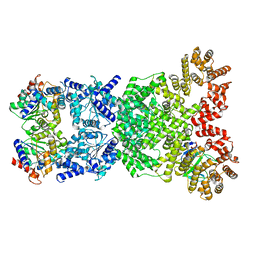 | |
8WYD
 
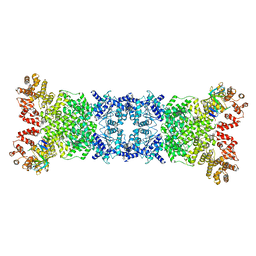 | | Cryo-EM structure of DSR2-DSAD1 complex | | 分子名称: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | 著者 | Zhang, J.T, Jia, N, Liu, X.Y. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYF
 
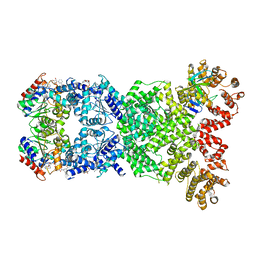 | |
8WY9
 
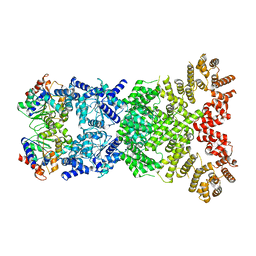 | |
8WYC
 
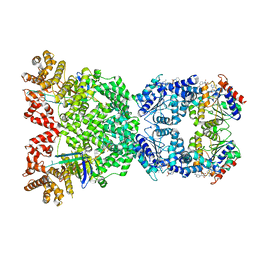 | |
8WYA
 
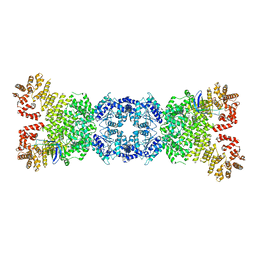 | | Cryo-EM structure of DSR2-tube complex | | 分子名称: | Bacillus phage SPbeta tube protein, SIR2 family protein | | 著者 | Zhang, J.T, Jia, N, Liu, X.Y. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYB
 
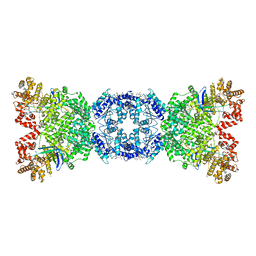 | | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ complex | | 分子名称: | Bacillus phage SPR Tube protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | 著者 | Zhang, J.T, Jia, N, Liu, X.Y. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WY8
 
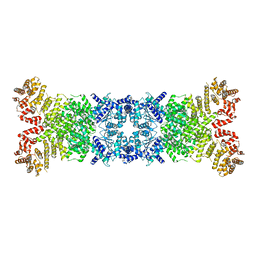 | |
5H9T
 
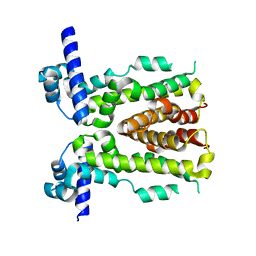 | | Crystal structure of native NalD at resolution of 2.9, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | 分子名称: | NalD | | 著者 | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | 登録日 | 2015-12-29 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa.
Mol. Microbiol., 100, 2016
|
|
7YOR
 
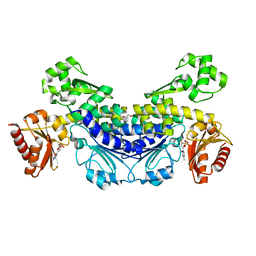 | | Recj2 and dCMP complex from Methanocaldococcus jannaschii | | 分子名称: | 1,2-ETHANEDIOL, [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-furan-2-yl]methyl dihydrogen phosphate, archaeal nuclease RecJ2 | | 著者 | Wang, W.W, Liu, X.P. | | 登録日 | 2022-08-02 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
4RL0
 
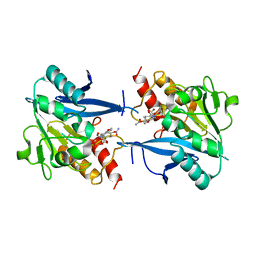 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | 分子名称: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | 著者 | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | 登録日 | 2014-10-14 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RL2
 
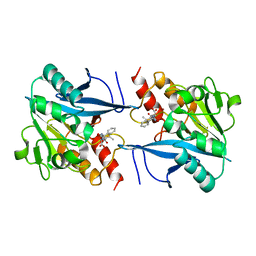 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | 分子名称: | (1R)-2-({(R)-carboxy[(2R,5S)-4-carboxy-5-methyl-5,6-dihydro-2H-1,3-thiazin-2-yl]methyl}amino)-2-oxo-1-phenylethanaminium, Beta-lactamase NDM-1, ZINC ION | | 著者 | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | 登録日 | 2014-10-15 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
1QFD
 
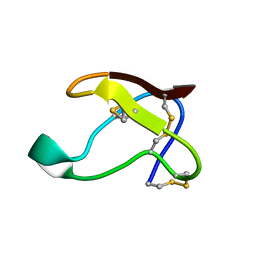 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | 分子名称: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | 著者 | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | 登録日 | 1999-04-08 | | 公開日 | 1999-07-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
1EJI
 
 | | RECOMBINANT SERINE HYDROXYMETHYLTRANSFERASE (MOUSE) | | 分子名称: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], SERINE HYDROXYMETHYLTRANSFERASE | | 著者 | Szebenyi, D.M.E, Liu, X, Kriksunov, I.A, Stover, P.J, Thiel, D.J. | | 登録日 | 2000-03-02 | | 公開日 | 2000-11-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers.
Biochemistry, 39, 2000
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | 分子名称: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-06-17 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1FJA
 
 | |
1FTN
 
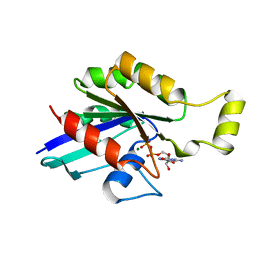 | | CRYSTAL STRUCTURE OF THE HUMAN RHOA/GDP COMPLEX | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA (H12) | | 著者 | Wei, Y, Zhang, Y, Derewenda, U, Liu, X, Minor, W, Nakamoto, R.K, Somlyo, A.V, Somlyo, A.P, Derewenda, Z.S. | | 登録日 | 1997-03-13 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of RhoA-GDP and its functional implications.
Nat.Struct.Biol., 4, 1997
|
|
1CZA
 
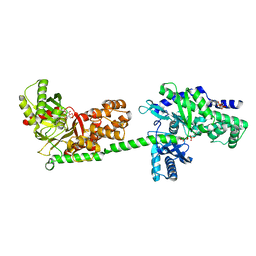 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I COMPLEXED WITH GLUCOSE, GLUCOSE-6-PHOSPHATE, AND ADP | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, ... | | 著者 | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | 登録日 | 1999-09-01 | | 公開日 | 2000-03-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
1DGK
 
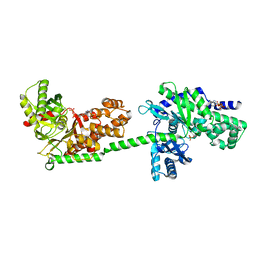 | | MUTANT MONOMER OF RECOMBINANT HUMAN HEXOKINASE TYPE I WITH GLUCOSE AND ADP IN THE ACTIVE SITE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, HEXOKINASE TYPE I, PHOSPHATE ION, ... | | 著者 | Aleshin, A.E, Liu, X, Kirby, C, Bourenkov, G.P, Bartunik, H.D, Fromm, H.J, Honzatko, R.B. | | 登録日 | 1999-11-24 | | 公開日 | 2000-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of mutant monomeric hexokinase I reveal multiple ADP binding sites and conformational changes relevant to allosteric regulation.
J.Mol.Biol., 296, 2000
|
|
3TOE
 
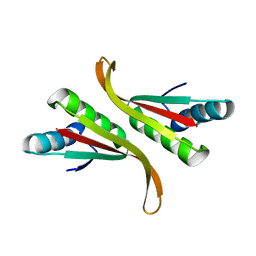 | | Structure of Mth10b | | 分子名称: | DNA/RNA-binding protein Alba | | 著者 | Pan, X.M, Zhang, N, Liu, Y.F, Liu, X. | | 登録日 | 2011-09-05 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | Molecular mechanism underlying the interaction of typical Sac10b family proteins with DNA.
Plos One, 7, 2012
|
|
