5YPM
 
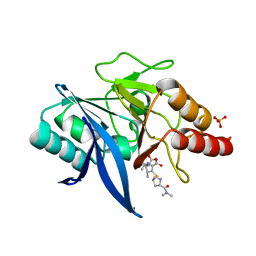 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
6K7T
 
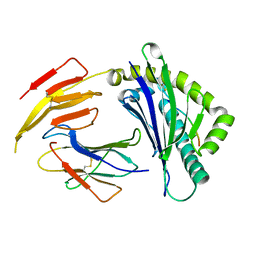 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1--human beta-2 microglobulin | | Descriptor: | Beta-2-microglobulin, HeV1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
7S71
 
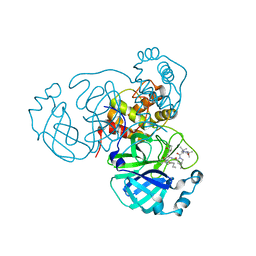 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI35 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(hexylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6X
 
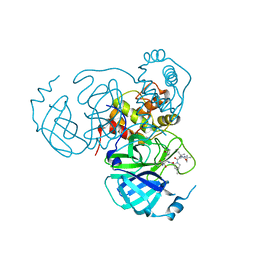 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI30 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S75
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI42 | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(3-methylbutanoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S72
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI36 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6W
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI29 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6Z
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S6Y
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI32 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-[(cyclopropylmethyl)amino]-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S74
 
 | |
7S73
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI37 | | Descriptor: | (6S)-5-{(2S)-2-[(tert-butylcarbamoyl)amino]-3,3-dimethylbutanoyl}-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-5-azaspiro[2.4]heptane-6-carboxamide (non-preferred name), 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7S70
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI34 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(butylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Yang, K.S, Sankaran, B, Liu, W.R. | | Deposit date: | 2021-09-15 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
3PBL
 
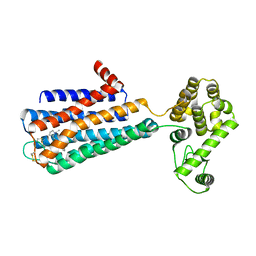 | | Structure of the human dopamine D3 receptor in complex with eticlopride | | Descriptor: | 3-chloro-5-ethyl-N-{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxy-2-methoxybenzamide, D(3) dopamine receptor, Lysozyme chimera, ... | | Authors: | Chien, E.Y.T, Liu, W, Han, G.W, Katritch, V, Zhao, Q, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of the human dopamine d3 receptor in complex with a d2/d3 selective antagonist.
Science, 330, 2010
|
|
6K7U
 
 | | Crystal structure of beta-2 microglobulin (beta2m) of Bat (Pteropus Alecto) | | Descriptor: | Bat beta-2-microglobulin | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
7KX0
 
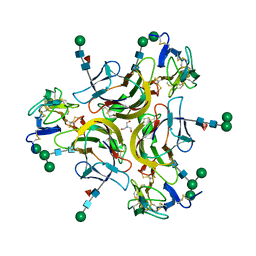 | | Crystal structure of the CD27:CD70 co-stimulatory complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD27 antigen, ... | | Authors: | Maben, Z, Liu, W, Mosyak, L, Chaparro-Riggers, J. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural delineation and phase-dependent activation of the costimulatory CD27:CD70 complex.
J.Biol.Chem., 297, 2021
|
|
5DTH
 
 | |
7JJO
 
 | | Structural Basis of the Activation of Heterotrimeric Gs-protein by Isoproterenol-bound Beta1-Adrenergic Receptor | | Descriptor: | Beta1-Adrenergic Receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Su, M, Zhu, L, Zhang, Y, Paknejad, N, Dey, R, Huang, J, Lee, M.Y, Williams, D, Jordan, K.D, Eng, E.T, Ernst, O.P, Meyerson, J.R, Hite, R.K, Walz, T, Liu, W, Huang, X.Y. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural Basis of the Activation of Heterotrimeric Gs-Protein by Isoproterenol-Bound beta 1 -Adrenergic Receptor.
Mol.Cell, 80, 2020
|
|
5DAB
 
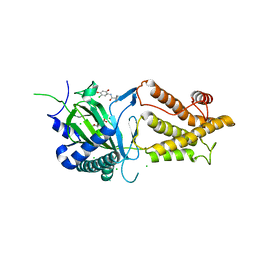 | | Crystal structure of FTO-IN115 | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase FTO, CHLORIDE ION, ... | | Authors: | Chai, J, Zhou, B, Liu, W, Han, Z, Niu, T. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of FTO-IN115
To Be Published
|
|
7TLL
 
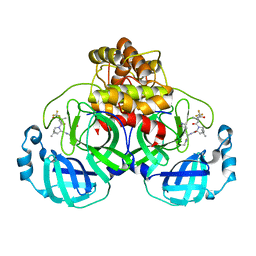 | | Structure of SARS-CoV-2 Mpro Omicron P132H in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-01-26 | | Last modified: | 2022-06-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
5IJ8
 
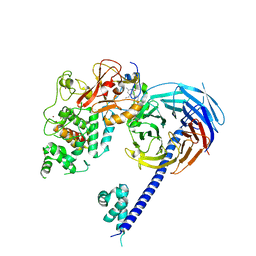 | | Structure of the primary oncogenic mutant Y641N Hs/AcPRC2 in complex with a pyridone inhibitor | | Descriptor: | 5,8-dichloro-2-[(4-ethyl-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-({1-[(2R)-2-hydroxypropanoyl]piperidin-4-yl}oxy)-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of Zeste Homolog 2 (EZH2),Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Deng, Y.-L, Liu, W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.
Nat Commun, 7, 2016
|
|
7LMC
 
 | |
2GLP
 
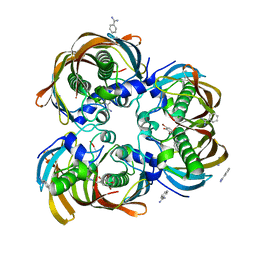 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with compound 1 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
5UVJ
 
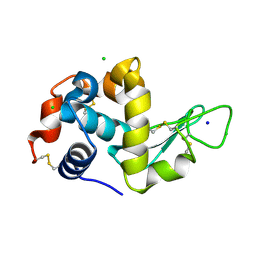 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
6L8Z
 
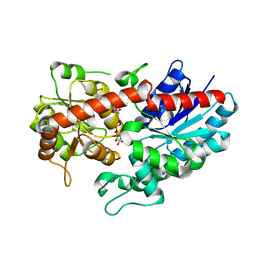 | | Crystal structure of ugt transferase mutant in complex with UPG | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2019-11-07 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
