1D3H
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH ANTIPROLIFERATIVE AGENT A771726 | | Descriptor: | (2Z)-2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]but-2-enamide, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Liu, S, Neidhardt, E.A, Grossman, T.H, Ocain, T, Clardy, J. | | Deposit date: | 1999-09-29 | | Release date: | 2000-08-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents.
Structure Fold.Des., 8, 2000
|
|
4LTY
 
 | | Crystal Structure of E.coli SbcD at 1.8 A Resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7X4R
 
 | |
6N7R
 
 | | Saccharomyces cerevisiae spliceosomal E complex (ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
5J95
 
 | | MAP4K4 in complex with inhibitor | | Descriptor: | 1-{4-[6-amino-5-(4-chlorophenyl)pyridin-3-yl]phenyl}cyclopentane-1-carboxylic acid, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Liu, S. | | Deposit date: | 2016-04-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MAP4K4 in complex with inhibitor
To Be Published
|
|
4DOY
 
 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
3JSI
 
 | | Human phosphodiesterase 9 in complex with inhibitor | | Descriptor: | 6-benzyl-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2009-09-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Identification of a Brain Penetrant PDE9A Inhibitor Utilizing Prospective Design and Chemical Enablement as a Rapid Lead Optimization Strategy.
J.Med.Chem., 52, 2009
|
|
3JSW
 
 | | Human PDE9 in complex with selective inhibitor | | Descriptor: | 6-[(3S,4S)-1-benzyl-4-methylpyrrolidin-3-yl]-1-(1-methylethyl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2009-09-11 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Brain Penetrant PDE9A Inhibitor Utilizing Prospective Design and Chemical Enablement as a Rapid Lead Optimization Strategy.
J.Med.Chem., 52, 2009
|
|
6LA4
 
 | | Cryo-EM structure of full echovirus 11 particle at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LA6
 
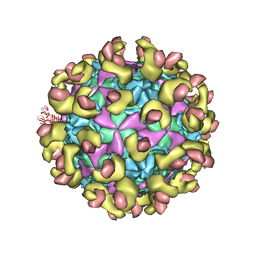 | |
6LAP
 
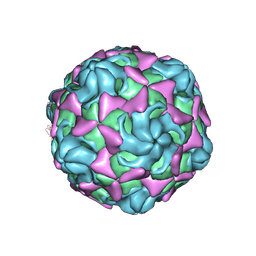 | | Cryo-EM structure of echovirus 11 A-particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
1M1B
 
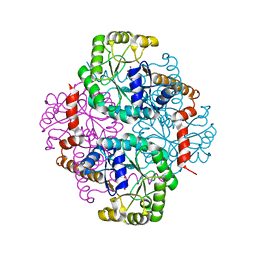 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | Authors: | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2002-06-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
1OQF
 
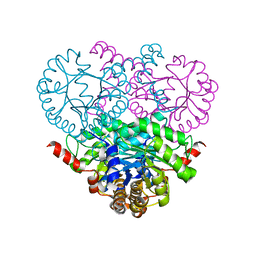 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
4LU9
 
 | | Crystal structure of E.coli SbcD at 2.5 angstrom resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M0V
 
 | | Crystal structure of E.coli SbcD with Mn2+ | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL, MANGANESE (II) ION | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YHV
 
 | | Yeast Prp3 C-terminal fragment 325-469 | | Descriptor: | ACETIC ACID, U4/U6 small nuclear ribonucleoprotein PRP3 | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
1BN5
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-07-31 | | Release date: | 1999-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
4YHW
 
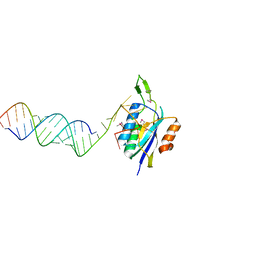 | | Yeast Prp3 (296-469) in complex with fragment of U4/U6 di-snRNA | | Descriptor: | U4 snRNA fragment, U4/U6 small nuclear ribonucleoprotein PRP3, U6 snRNA fragment | | Authors: | Liu, S, Wahl, M.C. | | Deposit date: | 2015-02-27 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing.
Elife, 4, 2015
|
|
1BOA
 
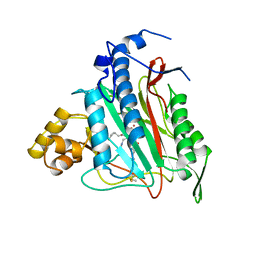 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH ANGIOGENESIS INHIBITOR FUMAGILLIN | | Descriptor: | COBALT (II) ION, FUMAGILLIN, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-08-01 | | Release date: | 1999-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
5W4W
 
 | |
1B6A
 
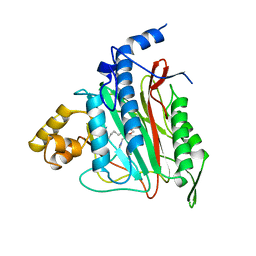 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH TNP-470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1B59
 
 | | COMPLEX OF HUMAN METHIONINE AMINOPEPTIDASE-2 COMPLEXED WITH OVALICIN | | Descriptor: | 3,4-DIHYDROXY-2-METHOXY-4-METHYL-3-[2-METHYL-3-(3-METHYL-BUT-2-ENYL) -OXIRANYL]-CYCLOHEXANONE, COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE) | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
3BLT
 
 | |
1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2U
 
 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
