8PO8
 
 | | Structure of Escherichia coli HrpA in complex with ADP and oligonucleotide poly(dC)11 forming an i-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO6
 
 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
2X22
 
 | | crystal structure of M. tuberculosis InhA inhibited by PT70 | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, DIMETHYL SULFOXIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], ... | | Authors: | Luckner, S.R, Liu, N, am Ende, C.W, Tonge, P.J, Kisker, C. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Slow, Tight Binding Inhibitor of Inha, the Enoyl-Acyl Carrier Protein Reductase from Mycobacterium Tuberculosis.
J.Biol.Chem., 285, 2010
|
|
2X23
 
 | | crystal structure of M. tuberculosis InhA inhibited by PT70 | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, DIMETHYL SULFOXIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], ... | | Authors: | Luckner, S.R, Liu, N, am Ende, C.W, Tonge, P.J, Kisker, C. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | A Slow, Tight Binding Inhibitor of Inha, the Enoyl-Acyl Carrier Protein Reductase from Mycobacterium Tuberculosis.
J.Biol.Chem., 285, 2010
|
|
5EX0
 
 | | Crystal structure of human SMYD3 in complex with a MAP3K2 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, MAP3K2 peptide, ... | | Authors: | Fu, W, Liu, N, Qiao, Q, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
4OIM
 
 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT119 in 2.4 M acetate | | Descriptor: | 2-(2-CYANOPHENOXY)-5-HEXYLPHENOL, ACETATE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-20 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Time-Dependent Diaryl Ether Inhibitors of InhA: Structure-Activity Relationship Studies of Enzyme Inhibition, Antibacterial Activity, and in vivo Efficacy.
Chemmedchem, 9, 2014
|
|
4OHU
 
 | | Crystal structure of Mycobacterium tuberculosis InhA in complex with inhibitor PT92 | | Descriptor: | 2-(2-bromophenoxy)-5-hexylphenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Yu, W, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
5EX3
 
 | | Crystal structure of human SMYD3 in complex with a VEGFR1 peptide | | Descriptor: | ACETIC ACID, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Qiao, Q, Fu, W, Liu, N, Wang, M, Min, J, Zhu, B, Xu, R.M, Yang, N. | | Deposit date: | 2015-11-23 | | Release date: | 2016-03-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Structural Basis for Substrate Preference of SMYD3, a SET Domain-containing Protein Lysine Methyltransferase
J.Biol.Chem., 291, 2016
|
|
5FTE
 
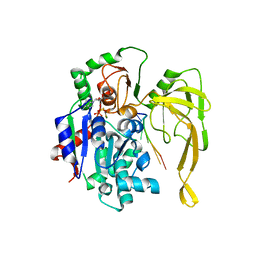 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP-AlF3 and ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTC
 
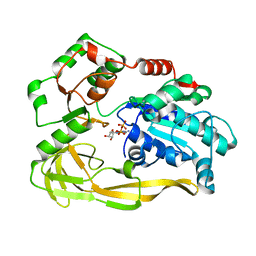 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTD
 
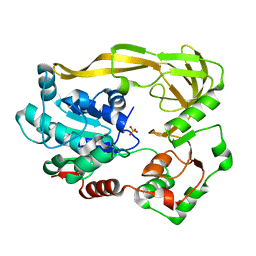 | | Crystal structure of Pif1 helicase from Bacteroides apo form | | Descriptor: | PHOSPHATE ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTF
 
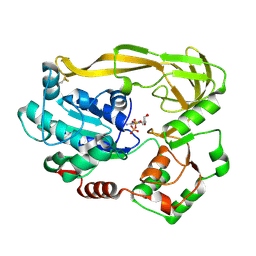 | | Crystal structure of Pif1 helicase from Bacteroides double mutant L95C-I339C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5G2N
 
 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
5FTB
 
 | | Crystal structure of Pif1 helicase from Bacteroides in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
4DYD
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-02-28 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E13
 
 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
4E12
 
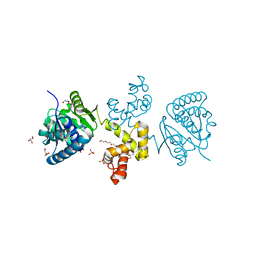 | | Substrate-directed dual catalysis of dicarbonyl compounds by diketoreductase | | Descriptor: | Diketoreductase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Lu, M, White, M.A, Huang, Y, Wu, X, Liu, N, Cheng, X, Chen, Y. | | Deposit date: | 2012-03-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dual catalysis mode for the dicarbonyl reduction catalyzed by diketoreductase
Chem.Commun.(Camb.), 48, 2012
|
|
3T88
 
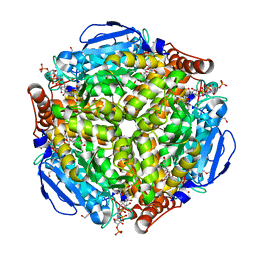 | | Crystal structure of Escherichia coli MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T8A
 
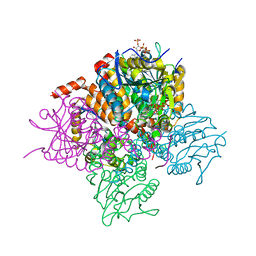 | | Crystal structure of Mycobacterium tuberculosis MenB in complex with substrate analogue, OSB-NCoA | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, o-succinylbenzoyl-N-coenzyme A | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T8B
 
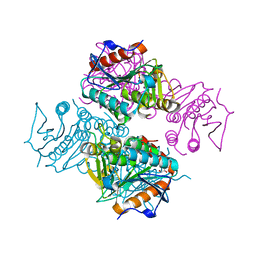 | | Crystal structure of Mycobacterium tuberculosis MenB with altered hexameric assembly | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T89
 
 | | Crystal structure of Escherichia coli MenB, the 1,4-dihydroxy-2-naphthoyl-CoA synthase in vitamin K2 biosynthesis | | Descriptor: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL, MALONATE ION | | Authors: | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
4HU8
 
 | | Crystal Structure of a Bacterial Ig-like Domain Containing GH10 Xylanase from Termite Gut | | Descriptor: | GH10 Xylanase, GLYCEROL, SULFATE ION | | Authors: | Han, Q, Liu, N, Robinson, H, Cao, L, Qian, C, Wang, Q, Xie, L, Ding, H, Wang, Q, Huang, Y, Li, J, Zhou, Z. | | Deposit date: | 2012-11-02 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical characterization and crystal structure of a GH10 xylanase from termite gut bacteria reveal a novel structural feature and significance of its bacterial Ig-like domain.
Biotechnol.Bioeng., 110, 2013
|
|
4OYR
 
 | | Competition of the small inhibitor PT91 with large fatty acyl substrate of the Mycobacterium tuberculosis enoyl-ACP reductase InhA by induced substrate-binding loop refolding | | Descriptor: | 2-(2-chloranylphenoxy)-5-hexyl-phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2995 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXY
 
 | | Substrate-binding loop movement with inhibitor PT10 in the tetrameric Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 5-hexyl-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Li, H.J, Sullivan, T.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-09 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3501 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
4OXN
 
 | | Substrate-like binding mode of inhibitor PT155 to the Mycobacterium tuberculosis enoyl-ACP reductase InhA | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ... | | Authors: | Li, H.J, Pan, P, Lai, C.T, Liu, N, Garcia-Diaz, M, Simmerling, C, Tonge, P.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2926 Å) | | Cite: | A Structural and Energetic Model for the Slow-Onset Inhibition of the Mycobacterium tuberculosis Enoyl-ACP Reductase InhA.
Acs Chem.Biol., 9, 2014
|
|
