5HII
 
 | | Crystal structure of glycine sarcosine N-methyltransferase (GSMT) from Methanohalophilus portucalensis (apo form) | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
5HIL
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylhomocysteine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
5HIK
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylmethionine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
3ROV
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2011-04-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
2GJ5
 
 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
3QI3
 
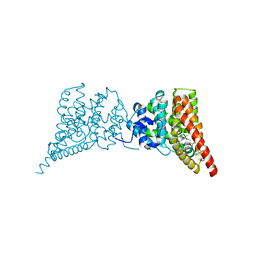 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
2H67
 
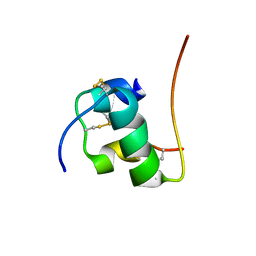 | | NMR structure of human insulin mutant HIS-B5-ALA, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Liu, M, Hu, S.Q, Jia, W, Arvan, P, Weiss, M.A. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | A Conserved Histidine in Insulin Is Required for the Foldability of Human Proinsulin: Structure and function of an Alab5 analog.
J.Biol.Chem., 281, 2006
|
|
3SFD
 
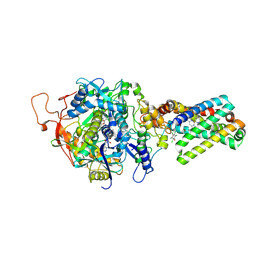 | | crystal structure of porcine mitochondrial respiratory complex II bound with oxaloacetate and pentachlorophenol | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhou, Q.J, Zhai, Y.J, Liu, M, Sun, F. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Protein Cell, 2, 2011
|
|
3SFE
 
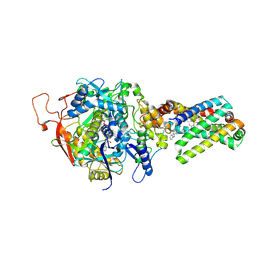 | | crystal structure of porcine mitochondrial respiratory complex II bound with oxaloacetate and thiabendazole | | Descriptor: | 2-(1,3-THIAZOL-4-YL)-1H-BENZIMIDAZOLE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Zhou, Q.J, Zhai, Y.J, Liu, M, Sun, F. | | Deposit date: | 2011-06-13 | | Release date: | 2011-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Thiabendazole inhibits ubiquinone reduction activity of mitochondrial respiratory complex II via a water molecule mediated binding feature.
Protein Cell, 2, 2011
|
|
3QI4
 
 | | Crystal structure of PDE9A(Q453E) in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
2KA9
 
 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | Descriptor: | Disks large homolog 4, cypin peptide | | Authors: | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
3PTJ
 
 | | Structural and functional Analysis of Arabidopsis thaliana thylakoid lumen protein AtTLP18.3 | | Descriptor: | UPF0603 protein At1g54780, chloroplastic | | Authors: | Wu, H.Y, Liu, M.S, Lin, T.P, Cheng, Y.S. | | Deposit date: | 2010-12-03 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen
Plant Physiol., 157, 2011
|
|
5GGM
 
 | | The NMR structure of calmodulin in CTAB reverse micelles | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
3PVH
 
 | | Structural and Functional Analysis of Arabidopsis thaliana thylakoid lumen protein AtTLP18.3 | | Descriptor: | CALCIUM ION, GLYCEROL, UPF0603 protein At1g54780, ... | | Authors: | Wu, H.Y, Liu, M.S, Lin, T.P, Cheng, Y.S. | | Deposit date: | 2010-12-07 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen
Plant Physiol., 157, 2011
|
|
7ENQ
 
 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
3PW9
 
 | | Structural and functional Analysis of Arabidopsis thaliana thylakoid lumen protein AtTLP18.3 | | Descriptor: | CALCIUM ION, GLYCEROL, SERINE, ... | | Authors: | Wu, H.Y, Liu, M.S, Lin, T.P, Cheng, Y.S. | | Deposit date: | 2010-12-08 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional assays of AtTLP18.3 identify its novel acid phosphatase activity in thylakoid lumen
Plant Physiol., 157, 2011
|
|
4HCC
 
 | | The zinc ion bound form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, ISOPROPYL ALCOHOL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
5FCG
 
 | |
4HCB
 
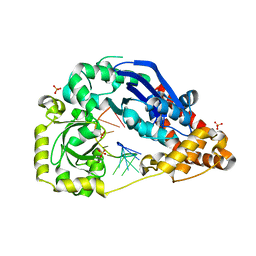 | | The metal-free form of crystal structure of E.coli ExoI-ssDNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), Exodeoxyribonuclease I, GLYCEROL, ... | | Authors: | Qiu, R, Wei, J, Lou, T, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2012-09-29 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of Escherichia coli exonuclease I in complex with the single strand DNA
To be published
|
|
2LHW
 
 | | Tri-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
2LI2
 
 | | Mono-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
3UZB
 
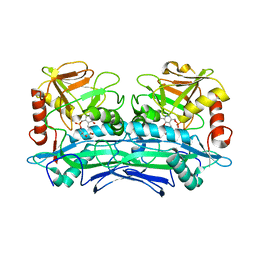 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
