7C7S
 
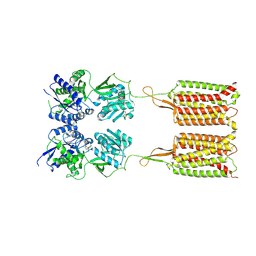 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | Descriptor: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | Authors: | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
7CKI
 
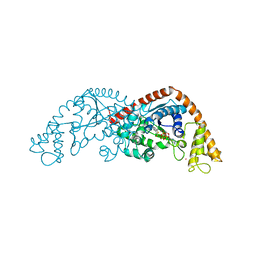 | | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP | | Descriptor: | (5~{S},6~{R})-5-methyl-7-thia-2-azatricyclo[6.3.1.0^{4,12}]dodeca-1(12),3,8,10-tetraene-6-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lyu, G, Fan, S, Jin, Y, Liu, J, Zou, S, Wu, G, Yang, Z. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Bacillus stearothermophilus in complex with chuangxinmycin and ATP
To Be Published
|
|
8ZB4
 
 | | Crystal structure of NudC from Mycobacterium abscessus in complex with NAD | | Descriptor: | CALCIUM ION, NAD(+) diphosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
8ZB5
 
 | | Crystal structure of NudC from Mycobacterium abscessus in complex with AMP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
8ZB3
 
 | | Crystal structure of NudC from Mycobacterium abscessus | | Descriptor: | CALCIUM ION, IMIDAZOLE, NAD(+) diphosphatase | | Authors: | Meng, L, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on Mycobacterial NudC Reveal a Class of Zinc Independent NADH Pyrophosphatase.
J.Mol.Biol., 436, 2024
|
|
4G43
 
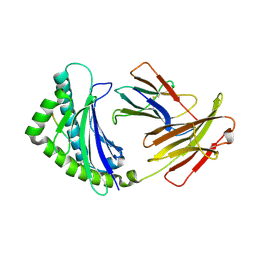 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
1LSL
 
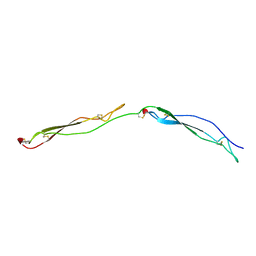 | | Crystal Structure of the Thrombospondin-1 Type 1 Repeats | | Descriptor: | Thrombospondin 1, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Tan, K, Duquette, M, Liu, J, Dong, Y, Zhang, R, Joachimiak, A, Lawler, J, Wang, J.-H. | | Deposit date: | 2002-05-17 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the TSP-1 type 1 repeats: a novel
layered fold and its biological implication.
J.Cell Biol., 159, 2002
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XES
 
 | | The structure of HLA-A/L1-1 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
6WN3
 
 | |
6WNB
 
 | |
6WNC
 
 | |
6WND
 
 | |
5WSH
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
8YZR
 
 | | The structure of HLA/peptide from virus | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1 | | Authors: | Min, L, Liu, J. | | Deposit date: | 2024-04-08 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Further cytotoxic T cell escapes of SARS-CoV-2 JN.1 explain its dominant circulation in recent waves
to be published
|
|
6VRV
 
 | | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model | | Descriptor: | 4-chloro-1-{(1S)-1-[(3S)-3-fluoropyrrolidin-3-yl]ethyl}-3-methyl-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Barberis, C.E, Batchelor, J.D, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6VRU
 
 | | PIM-inhibitor complex 1 | | Descriptor: | 3,4-dichloro-2-cyclopropyl-1-[(piperidin-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Barberis, C.E, Batchelor, J.D, Mechin, I, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5Y46
 
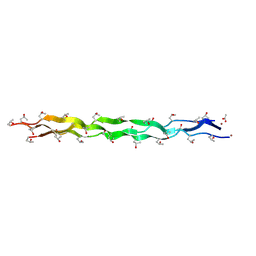 | |
8H59
 
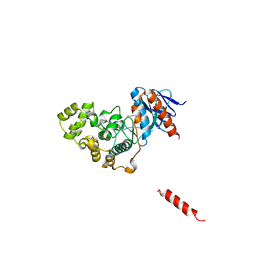 | | A fungal MAP kinase in complex with an inhibitor | | Descriptor: | Mitogen-activated protein kinase MPS1, ~{N}-[(2~{S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]-8-[2-methoxy-5-(trifluoromethyloxy)phenyl]-1,6-naphthyridine-2-carboxamide | | Authors: | Kong, Z, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi.
Mbio, 14, 2023
|
|
2W4V
 
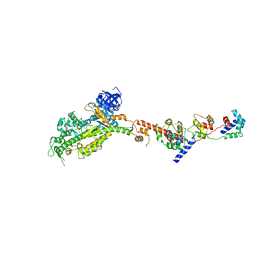 | | Isometrically contracting insect asynchronous flight muscle quick frozen after a quick release step | | Descriptor: | MYOSIN ESSENTIAL LIGHT CHAIN, STRIATED ADDUCTOR MUSCLE, MYOSIN HEAVY CHAIN, ... | | Authors: | Wu, S, Liu, J, Reedy, M.C, Tregear, R.T, Winkler, H, Franzini-Armstrong, C, Sasaki, H, Lucaveche, C, Goldman, Y.E, Reedy, M.K, Taylor, K.A. | | Deposit date: | 2008-12-02 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Structural Changes in Isometrically Contracting Insect Flight Muscle Trapped Following a Mechanical Perturbation.
Plos One, 7, 2012
|
|
4HSV
 
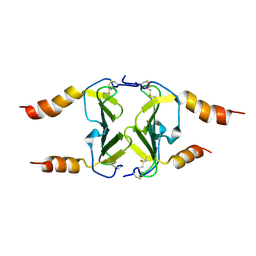 | | Crystal Structure of CXCL4L1 | | Descriptor: | Platelet factor 4 variant | | Authors: | Kuo, J.H, Liu, J.S, Wu, W.G, Sue, S.C. | | Deposit date: | 2012-10-30 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Alternative C-terminal helix orientation alters chemokine function: structure of the anti-angiogenic chemokine, CXCL4L1
J.Biol.Chem., 288, 2013
|
|
3MZ8
 
 | | Crystal Structure of Zinc-Bound Natrin From Naja atra | | Descriptor: | Natrin-1, ZINC ION | | Authors: | Wang, Y.L, Hsieh, Y.C, Liu, J.S, Chen, C.J, Wu, W.G. | | Deposit date: | 2010-05-12 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cobra CRISP functions as an inflammatory modulator via a novel Zn2+- and heparan sulfate- dependent transcriptional regulation of endothelial cell adhesion molecules
J.Biol.Chem., 285, 2010
|
|
