6O1O
 
 | |
5G06
 
 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
6NY3
 
 | | CasX ternary complex with 30bp target DNA | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6NY2
 
 | | CasX-gRNA-DNA(45bp) state I | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6NY1
 
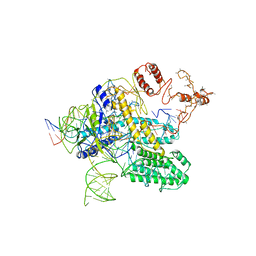 | | CasX-gRNA-DNA(30bp) State II | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
8IYQ
 
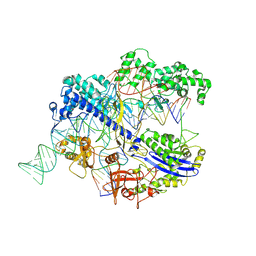 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8K0S
 
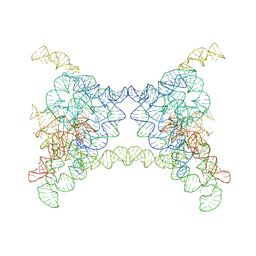 | |
8K15
 
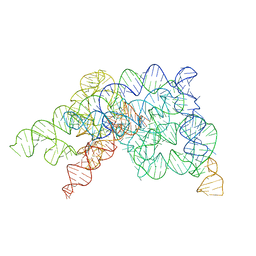 | |
8K0R
 
 | |
8K0Q
 
 | |
8K0P
 
 | |
7YOJ
 
 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
6P7M
 
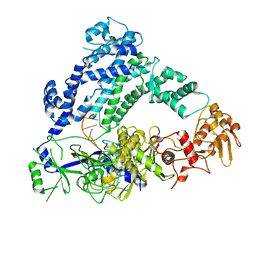 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (1:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
6P7N
 
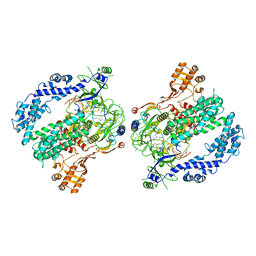 | | Cryo-EM structure of LbCas12a-crRNA: AcrVA4 (2:2 complex) | | Descriptor: | Cas12a, MAGNESIUM ION, anti-CRISPR VA4, ... | | Authors: | Knott, G.J, Liu, J.J, Doudna, J.A. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a.
Elife, 8, 2019
|
|
8Y2I
 
 | |
8XYC
 
 | |
3LE6
 
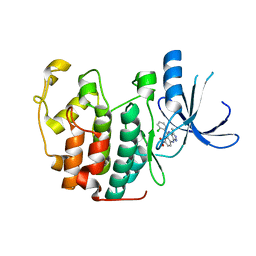 | | The structure of cyclin dependent kinase 2 (CKD2) with a pyrazolobenzodiazepine inhibitor | | Descriptor: | 5-(2-chlorophenyl)-3-methyl-7-nitropyrazolo[3,4-b][1,4]benzodiazepine, Cell division protein kinase 2 | | Authors: | Lukacs, C.M, Swain, A, Crowther, R.L, Kammlott, R.U, Liu, J.J. | | Deposit date: | 2010-01-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolobenzodiazepines: part I. Synthesis and SAR of a potent class of kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6MCB
 
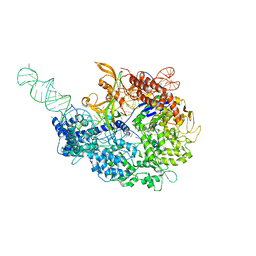 | |
5VZL
 
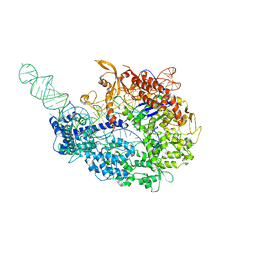 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
8IBX
 
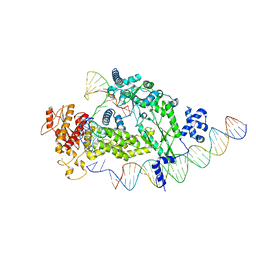 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
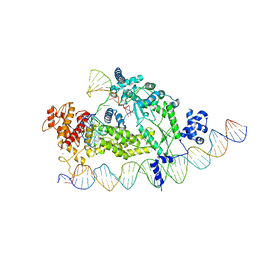 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
 | | Structure of R2 with 5'ORF and 3'UTR | | Descriptor: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBY
 
 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
5WFE
 
 | | Cas1-Cas2-IHF-DNA holo-complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Wright, A.V, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
3U7D
 
 | | Crystal structure of the KRIT1/CCM1 FERM domain in complex with the heart of glass (HEG1) cytoplasmic tail | | Descriptor: | Krev interaction trapped protein 1, Protein HEG homolog 1 | | Authors: | Gingras, A.R, Liu, J.J, Ginsberg, M.H, Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2011-10-13 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of the junctional anchorage of the cerebral cavernous malformations complex.
J.Cell Biol., 199, 2012
|
|
