8W4R
 
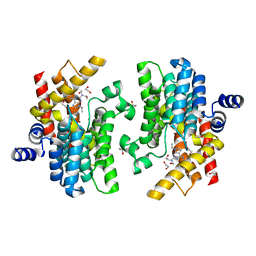 | | Crystal structure of PDE4D complexed with CVT-313 | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4T
 
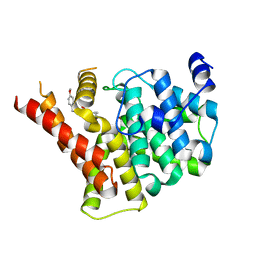 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | 2-[bis(2-hydroxyethyl)amino]-6-[(4-methoxyphenyl)methylamino]-9-propan-2-yl-7~{H}-purin-8-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8WDO
 
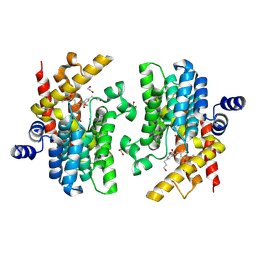 | | Crystal structure of PDE4D complexed with DCN | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-(3,4-dimethoxyphenyl)-3-(hydroxymethyl)-7-methoxy-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
8WDN
 
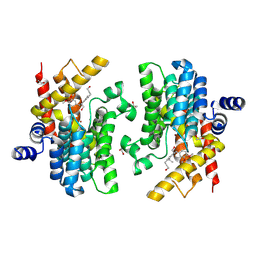 | | Crystal structure of PDE4D complexed with 7b-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-7-ethoxy-2-(3-ethoxy-4-methoxy-phenyl)-3-(hydroxymethyl)-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
3EQR
 
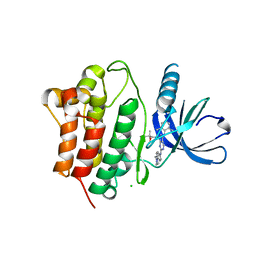 | | Crystal Structure of Ack1 with compound T74 | | Descriptor: | Activated CDC42 kinase 1, CHLORIDE ION, N~3~-(2,6-dimethylphenyl)-1-(3-methoxy-3-methylbutyl)-N~6~-(4-piperazin-1-ylphenyl)-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamine | | Authors: | Liu, J, Wang, Z, Walker, N.P.C. | | Deposit date: | 2008-10-01 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of N3,N6-diaryl-1H-pyrazolo[3,4-d]pyrimidine-3,6-diamines as a novel class of ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7RGW
 
 | |
3EEK
 
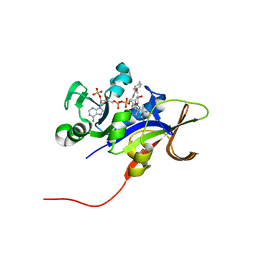 | |
3EEM
 
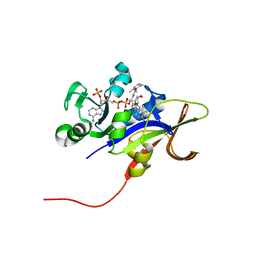 | | Candida glabrata Dihydrofolate Reductase complexed with 2,4-diamino-5-[3-methyl-3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)prop-1-ynyl]-6-methylpyrimidine(UCP111D26M) and NADPH | | Descriptor: | 5-[(3R)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors
Chem.Biol.Drug Des., 73, 2009
|
|
3EEL
 
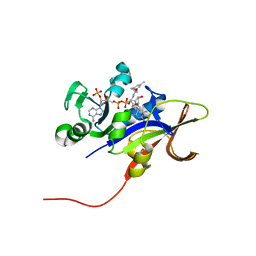 | | Candida glabrata Dihydrofolate Reductase complexed with 2,4-diamino-5-[3-methyl-3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)prop-1-ynyl]-6-methylpyrimidine(UCP11153TM) and NADPH | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors
Chem.Biol.Drug Des., 73, 2009
|
|
3EEJ
 
 | |
3CK4
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, MAGNESIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
7SC4
 
 | | filamin complex-1 | | Descriptor: | Filamin-A/Integrin alpha-IIb light chain, form 2 chimera, SULFATE ION | | Authors: | Liu, J, Qin, J. | | Deposit date: | 2021-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A mechanism of platelet integrin alpha IIb beta 3 outside-in signaling through a novel integrin alpha IIb subunit-filamin-actin linkage.
Blood, 141, 2023
|
|
7SFT
 
 | | Filamin complex-2 | | Descriptor: | Filamin-A, Integrin alpha-IIb light chain, form 2 | | Authors: | Liu, J, Qin, J. | | Deposit date: | 2021-10-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A mechanism of platelet integrin alpha IIb beta 3 outside-in signaling through a novel integrin alpha IIb subunit-filamin-actin linkage.
Blood, 141, 2023
|
|
3CSE
 
 | | Candida glabrata Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A.C. | | Deposit date: | 2008-04-09 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided development of efficacious antifungal agents targeting Candida glabrata dihydrofolate reductase.
Chem.Biol., 15, 2008
|
|
8I5Z
 
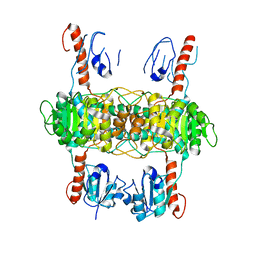 | |
3CRP
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-04-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
5F1I
 
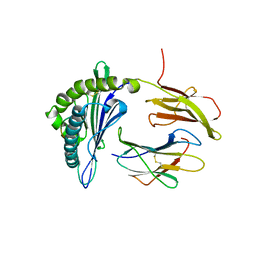 | | MHC with 9-mer peptide | | Descriptor: | 9-mer peptide, Beta2M, MHC class I DLA-88 | | Authors: | Liu, J, Chai, Y, QI, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Diversified Anchoring Features the Peptide Presentation of DLA-88*50801: First Structural Insight into Domestic Dog MHC Class I
J Immunol., 197, 2016
|
|
5WUW
 
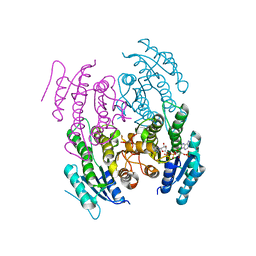 | |
5WEH
 
 | | Cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Aa3-type cytochrome c oxidase subunit IV, CALCIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, F, Ling, Q, Hiser, C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity.
Biochem. Soc. Trans., 45, 2017
|
|
5F1N
 
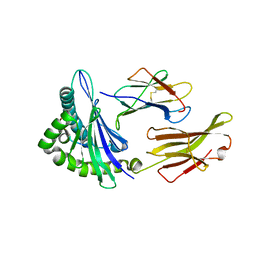 | | MHC complexed to 11mer peptide | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Peptide from Cytochrome P450 family 1 subfamily B polypeptide 1 | | Authors: | Liu, J, Chai, Y, Qi, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversified Anchoring Features the Peptide Presentation of DLA-88*50801: First Structural Insight into Domestic Dog MHC Class I
J Immunol., 197, 2016
|
|
3G9R
 
 | |
7RAM
 
 | | Cryo-EM Structure of the HCMV gHgLgO Trimer Derived from AD169 and TR strains in complex with PDGFRalpha | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, Envelope glycoprotein O, ... | | Authors: | Liu, J, Vanarsdall, A.L, Chen, D, Johnson, D.C, Jardetzky, T.S. | | Deposit date: | 2021-07-02 | | Release date: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-Electron Microscopy Structure and Interactions of the Human Cytomegalovirus gHgLgO Trimer with Platelet-Derived Growth Factor Receptor Alpha.
Mbio, 12, 2021
|
|
6K0Y
 
 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
6CI0
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with E101A (II) mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, ... | | Authors: | Liu, J, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2018-02-23 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The K-path entrance in cytochrome c oxidase is defined by mutation of E101 and controlled by an adjacent ligand binding domain.
Biochim. Biophys. Acta, 1859, 2018
|
|
4NT6
 
 | | HLA-C*0801 Crystal Structure | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, Cw-8 alpha chain, ... | | Authors: | Liu, J.X, Toh, X.Y, Ren, E.C. | | Deposit date: | 2013-12-01 | | Release date: | 2014-07-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans.
J.Virol., 88, 2014
|
|
