4OHR
 
 | | Crystal structure of MilB from Streptomyces rimofaciens | | Descriptor: | CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
4OHB
 
 | | Crystal structure of MilB E103A in complex with 5-hydroxymethylcytidine 5'-monophosphate (hmCMP) from Streptomyces rimofaciens | | Descriptor: | 5-(hydroxymethyl)cytidine 5'-(dihydrogen phosphate), CMP/hydroxymethyl CMP hydrolase | | Authors: | Zhao, G, Zhang, Y, Liu, G, Wu, G, He, X. | | Deposit date: | 2014-01-17 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the N-glycosidase MilB in complex with hydroxymethyl CMP reveals its Arg23 specifically recognizes the substrate and controls its entry
Nucleic Acids Res., 42, 2014
|
|
5TTT
 
 | | Sparse-restraint solution NMR structure of micelle-solubilized cytosolic amino terminal domain of C. elegans mechanosensory ion channel MEC-4 refined by restrained Rosetta | | Descriptor: | Degenerin mec-4 | | Authors: | Everett, J.K, Liu, G, Mao, B, Driscoll, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparse-restraint solution NMR structure of micelle-solubilized
cytosolic amino terminal domain of C. elegans mechanosensory ion channel
MEC-4 refined by restrained Rosetta
To Be Published
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
7WBO
 
 | | Crystal structure of Sarcoplasmic Calcium-Binding Protein from Scylla paramamosain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Chen, Y, Jin, T, Liu, G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure Analysis of Sarcoplasmic-Calcium-Binding Protein: An Allergen in Scylla paramamosain.
J.Agric.Food Chem., 71, 2023
|
|
2FGX
 
 | | Solution NMR Structure of Protein Ne2328 from Nitrosomonas europaea. Northeast Structural Genomics Consortium Target NeT3. | | Descriptor: | putative thioredoxin | | Authors: | Eletsky, A, Liu, G, Yee, A, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-22 | | Release date: | 2006-02-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Nitrosomonas Europaea Hypothetical Protein Ne2328: Northeast Structural Genomics Consortium Target NeT3
To be Published
|
|
2HFV
 
 | | Solution NMR Structure of Protein RPA1041 from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium Target PaT90. | | Descriptor: | Hypothetical Protein RPA1041 | | Authors: | Eletsky, A, Atreya, H.S, Liu, G, Sukumaran, D, Garcia, M, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Pseudomonas aeruginosa Hypothetical Protein RPA1041
TO BE PUBLISHED
|
|
2JNA
 
 | | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 | | Descriptor: | Putative secreted protein | | Authors: | Eletsky, A, Parish, D, Liu, G, Sukumaran, D, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-31 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109
To be Published
|
|
2K2B
 
 | |
2JZT
 
 | | Solution NMR structure of Q8ZP25_SALTY from Salmonella typhimurium. Northeast Structural Genomics Consortium target StR70 | | Descriptor: | Putative thiol-disulfide isomerase and thioredoxin | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T, Bansal, S, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
2GZP
 
 | | Solution NMR structure of Q8ZP25 from Salmonella typhimurium LT2; Northeast Structural Genomics Consortium Target STR70 | | Descriptor: | putative chaperone | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Q8ZP24 from Salmonella typhimurium LT2.
To be Published
|
|
2GZO
 
 | | NMR structure of UPF0301 PROTEIN SO3346 from Shewanella oneidensis: Northeast Structural Genomics Consortium target SOR39 | | Descriptor: | UPF0301 protein SO3346 | | Authors: | Singarapu, K.K, Liu, G, Eletsky, A, Xu, D, Sukumaran, D.K, Mei, J, Xiao, R, Cunningham, K, Ma, L.C, Ritu, S, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of UPF0301 PROTEIN SO3346 from Shewanella oneidensis: Northeast Structural Genomics Consortium target SOR39
To be Published
|
|
2H3J
 
 | | Solution NMR Structure of Protein PA4359 from Pseudomonas aeruginosa: Northeast Structural Genomics Consortium Target PaT89 | | Descriptor: | Hypothetical protein PA4359 | | Authors: | Zhang, Q, Liu, G, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hypothetical protein PA4359: Northest Structural Genomics Target PaT89
To be Published
|
|
2MA4
 
 | | Solution NMR Structure of yahO protein from Salmonella typhimurium, Northeast Structural Genomics Consortium (NESG) Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Eletsky, A, Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
2LNI
 
 | | Solution NMR Structure of Stress-induced-phosphoprotein 1 STI1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4403E | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Tang, Y, Liu, G, Hamilton, K, Ciccosanti, C, Shastry, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4403E
To be Published
|
|
2M89
 
 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
8WRB
 
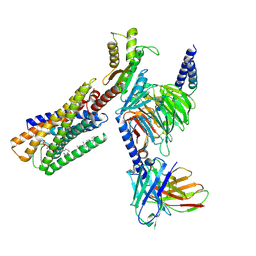 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
8XJK
 
 | | Cloprosetnol bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJO
 
 | | U46619 bound Thromboxane A2 receptor-Gq Protein Complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJN
 
 | | Cloprosetnol bound Thromboxane A2 receptor-Gq Protein Complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJL
 
 | | PGF2-alpha bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJM
 
 | | Latanoprost acid bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | Antibody fragment scFv16, Engineered miniGq, Fusion tag,Prostaglandin F2-alpha receptor,LgBiT, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
7CC9
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*GP*CP*GP*GS*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*CP*CP*GP*CP*C)-3'), ... | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCJ
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*AP*TP*C)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Zhao, G, Gan, J, Liu, G, Wu, G, He, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CCD
 
 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
