4M1J
 
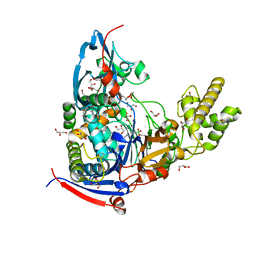 | | Crystal structure of Pseudomonas aeruginosa PvdQ in complex with a transition state analogue | | Descriptor: | Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, GLYCEROL, ... | | Authors: | Wu, R, Clevenger, K, Er, J, Fast, W.L, Liu, D. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of a Transition State Analogue with Picomolar Affinity for Pseudomonas aeruginosa PvdQ, a Siderophore Biosynthetic Enzyme.
Acs Chem.Biol., 8, 2013
|
|
4FM4
 
 | | Wild Type Fe-type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit, ... | | Authors: | Kuhn, M.L, Martinez, S, Gumataotao, N, Bornscheuer, U, Liu, D, Holz, R.C. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | The Fe-type nitrile hydratase from Comamonas testosteroni Ni1 does not require an activator accessory protein for expression in Escherichia coli.
Biochem.Biophys.Res.Commun., 424, 2012
|
|
3RZ2
 
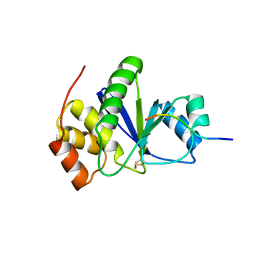 | | Crystal of Prl-1 complexed with peptide | | Descriptor: | Prl-1 (PTP4A1), Protein tyrosine phosphatase type IVA 1 | | Authors: | Zhang, Z.-Y, Liu, D, Bai, Y. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-26 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein.
J.Biol.Chem., 286, 2011
|
|
4PVG
 
 | |
4RBN
 
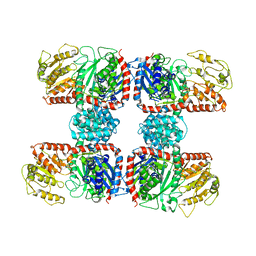 | | The crystal structure of Nitrosomonas europaea sucrose synthase: Insights into the evolutionary origin of sucrose metabolism in prokaryotes | | Descriptor: | Sucrose synthase:Glycosyl transferases group 1 | | Authors: | Wu, R, Asencion Diez, M.D, Figueroa, C.M, Machtey, M, Iglesias, A.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
J.Bacteriol., 197, 2015
|
|
6J8I
 
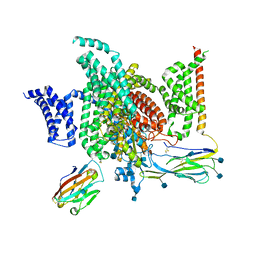 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 up) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
6J8G
 
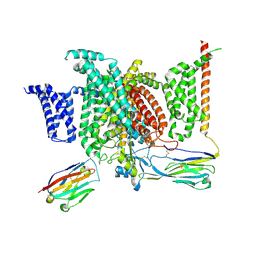 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (Y1755 up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 9 subunit alpha, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
6J8H
 
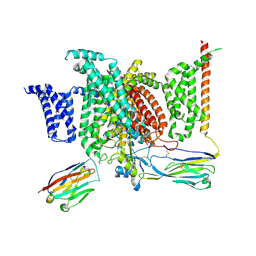 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (Y1755 down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 9 subunit alpha, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
6J8J
 
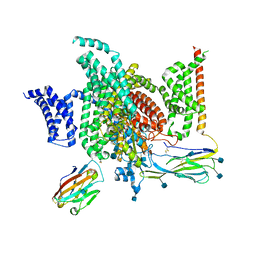 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 down) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
7EZO
 
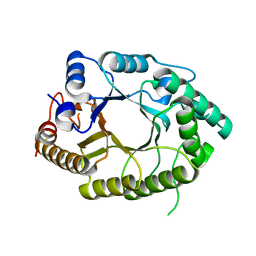 | |
7LJT
 
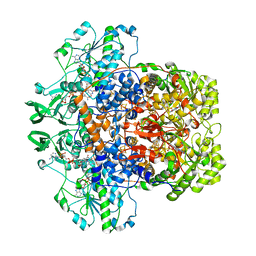 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) soaked with 5-Ethynyluracil (5EU), NADPH - 20 minutes | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LJS
 
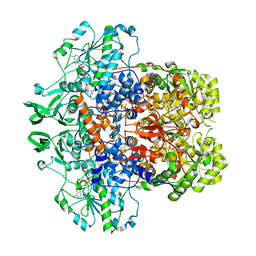 | | Porcine Dihydropyrimidine dehydrogenase (DPD) complexed with 5-Ethynyluracil (5EU) - Open Form | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LJU
 
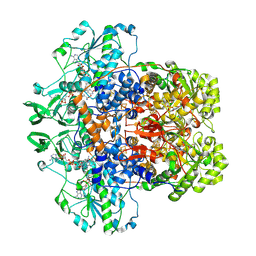 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) crosslinked with 5-Ethynyluracil (5EU) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LNM
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclopentene-1-carboxylic acid | | Descriptor: | (1~{R},3~{S},4~{R})-3-methyl-4-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Catlin, D, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7L5F
 
 | |
7JH4
 
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
7JX9
 
 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
7LON
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (1R,3S,4R)-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-methylcyclohexane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LOM
 
 | | Ornithine Aminotransferase (OAT) soaked with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (3~{S},4~{S})-4-methyl-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, (4~{R})-4-(fluoranylmethyl)-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
2K6B
 
 | |
7W26
 
 | | monolignol ferulate transferase | | Descriptor: | Ferulate monolignol transferase | | Authors: | Xi, L, Shuliu, D, Yue, F, Yi, Z. | | Deposit date: | 2021-11-22 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structure of the plant feruloyl-coenzyme A monolignol transferase provides insights into the formation of monolignol ferulate conjugates.
Biochem.Biophys.Res.Commun., 594, 2022
|
|
6BQR
 
 | | Human TRPM4 ion channel in lipid nanodiscs in a calcium-free state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Autzen, H.E, Myasnikov, A.G, Campbell, M.G, Asarnow, D, Julius, D, Cheng, Y. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science, 359, 2018
|
|
6BQV
 
 | | Human TRPM4 ion channel in lipid nanodiscs in a calcium-bound state | | Descriptor: | CALCIUM ION, CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Autzen, H.E, Myasnikov, A.G, Campbell, M.G, Asarnow, D, Julius, D, Cheng, Y. | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human TRPM4 ion channel in a lipid nanodisc.
Science, 359, 2018
|
|
6O20
 
 | | Cryo-EM structure of TRPV5 with calmodulin bound | | Descriptor: | CALCIUM ION, Calmodulin, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O1N
 
 | | Cryo-EM structure of TRPV5 (1-660) in nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 5 | | Authors: | Dang, S, van Goor, M.K, Asarnow, D, Wang, Y, Julius, D, Cheng, Y, van der Wijst, J. | | Deposit date: | 2019-02-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insight into TRPV5 channel function and modulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
