5UGQ
 
 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
4ZSY
 
 | | Pig Brain GABA-AT inactivated by (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic acid. | | Descriptor: | (1S)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-3-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Wu, R, Lee, H, Le, H.V, Doud, E, Sanishvili, R, Compton, P, Kelleher, N.L, Silverman, R.B, Liu, D. | | Deposit date: | 2015-05-14 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem.Biol., 10, 2015
|
|
8F6N
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine Quasi-Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
5UOH
 
 | | Crystal Structure of Hip1 (Rv2224c) T466A mutant | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Baikovitz, J, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5T4K
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4L
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UBK
 
 | | Inactive S1A/N269D-cpPvdQ mutant in complex with the pyoverdine precursor PVDIq reveals a specific binding pocket for the D-Tyr of this substrate | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, N-[(1R)-1-{(6S)-6-[(2-amino-2-oxoethyl)carbamoyl]-1,4,5,6-tetrahydropyrimidin-2-yl}-2-(4-hydroxyphenyl)ethyl]-N~2~-tetradecanoyl-L-glutamine | | Authors: | Mascarenhas, R, Catlin, D, Wu, R, Clevenger, K, Fast, W, Liu, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-01 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Circular Permutation Reveals a Chromophore Precursor Binding Pocket of the Siderophore Tailoring Enzyme PvdQ
To Be Published
|
|
4PVG
 
 | |
5UNO
 
 | | Crystal Structure of Hip1 (Rv2224c) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5BTR
 
 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
3RZ2
 
 | | Crystal of Prl-1 complexed with peptide | | Descriptor: | Prl-1 (PTP4A1), Protein tyrosine phosphatase type IVA 1 | | Authors: | Zhang, Z.-Y, Liu, D, Bai, Y. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-26 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein.
J.Biol.Chem., 286, 2011
|
|
7S5I
 
 | | Crystal structure of Aldose-6-phosphate reductase (Ald6PRase) from peach (Prunus persica) leaves | | Descriptor: | Sorbitol-6-phosphate dehydrogenase | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
5VWO
 
 | | Ornithine aminotransferase inactivated by (1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | Descriptor: | (1S,3S,4E)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
6BYZ
 
 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocyclic Inhibitor 37 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALA-ALA-ALA, Insulin-degrading enzyme, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95624852 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
5W5R
 
 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase P96A mutant bound to activator pyruvate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, PYRUVIC ACID, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5VWR
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP)-alpha-ketoglutarate | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, Aspartate aminotransferase, GLYCEROL | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
7S5F
 
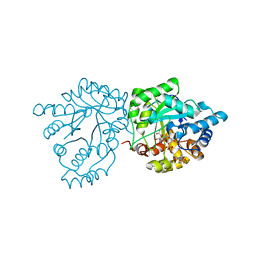 | | Crystal structure of mannose-6-phosphate reductase from celery (Apium graveolens) leaves with NADP+ and mannonic acid bound | | Descriptor: | D-MANNONIC ACID, Manose-6-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
5VWQ
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase | | Authors: | Mascarenhas, R, Lehrer, H, Liu, D, Ringe, D. | | Deposit date: | 2017-05-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
5W6J
 
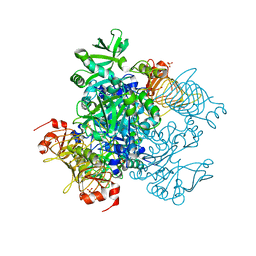 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Wu, R, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
7YBU
 
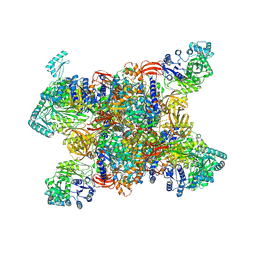 | | Human propionyl-coenzyme A carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Su, J.Y, Liu, D.S. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human propionyl-coenzyme A carboxylase
To Be Published
|
|
4IRW
 
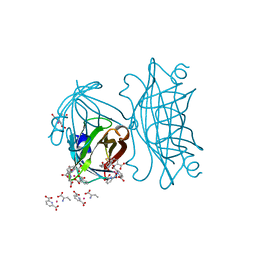 | | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form | | Descriptor: | BIOTIN, PYRIDINE-2,6-DICARBOXYLIC ACID, SODIUM ION, ... | | Authors: | Bandara, R.A.M.S.S, Liu, D.Q, Hindupur, A, Tesh, K.F, Fox, R.O. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form
To be Published
|
|
8V9M
 
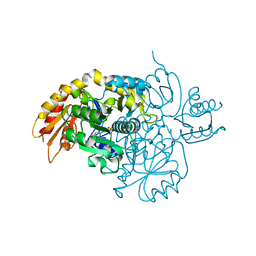 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (R)-3-amino-5,5-difluorocyclohex-1-ene-1-carboxylic acid. | | Descriptor: | 3-fluoro-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]benzoic acid, GLYCEROL, Ornithine aminotransferase, ... | | Authors: | Vargas, A.L, Devitt, A, Kaley, N, Silverman, R, Liu, D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, and Mechanistic Studies of ( R )-3-Amino-5,5-difluorocyclohex-1-ene-1-carboxylic Acid as an Inactivator of Human Ornithine Aminotransferase.
Acs Chem.Biol., 19, 2024
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
3U4W
 
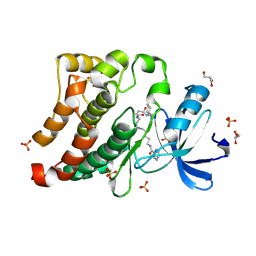 | | Src in complex with DNA-templated macrocyclic inhibitor MC4b | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION, ... | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
